

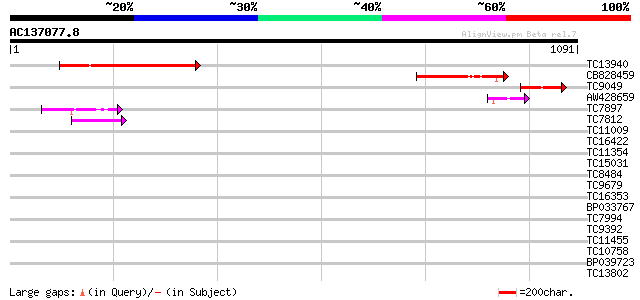
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137077.8 - phase: 0
(1091 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC13940 380 e-106
CB828459 231 6e-61
TC9049 similar to UP|O23575 (O23575) G2484-1 protein (Fragment),... 112 2e-25
AW428659 69 4e-12
TC7897 weakly similar to UP|ENL2_ARATH (Q9T076) Early nodulin-li... 50 3e-06
TC7812 UP|Q9LKJ6 (Q9LKJ6) Water-selective transport intrinsic me... 42 7e-04
TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Ar... 41 0.001
TC16422 similar to PIR|T05653|T05653 amino acid transport protei... 39 0.003
TC11354 similar to UP|CAE54480 (CAE54480) Alpha glucosidase II ... 39 0.004
TC15031 weakly similar to UP|Q69107 (Q69107) LAT protein, partia... 37 0.013
TC8484 similar to UP|Q9XGP1 (Q9XGP1) ESTs AU070372(S13446), part... 37 0.017
TC9679 weakly similar to UP|Q8H1B5 (Q8H1B5) Hin1-like protein, p... 35 0.064
TC16353 weakly similar to UP|HQGT_ARATH (Q9M156) Probable hydroq... 35 0.084
BP033767 35 0.084
TC7994 similar to UP|Q8RU73 (Q8RU73) Ribose-5-phosphate isomeras... 35 0.084
TC9392 35 0.084
TC11455 similar to UP|Q9ZUS8 (Q9ZUS8) At2g37380 protein, partial... 34 0.14
TC10758 34 0.14
BP039723 34 0.14
TC13802 similar to UP|Q9M1T1 (Q9M1T1) Sugar-phosphate isomerase-... 34 0.14
>TC13940
Length = 808
Score = 380 bits (976), Expect = e-106
Identities = 203/271 (74%), Positives = 225/271 (82%), Gaps = 1/271 (0%)
Frame = -1
Query: 97 LQSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNN 156
LQSS L RGSVVDY Q L LHPYQSP RNFLGH+TSW+SQA +RGPWI SPTPAP +
Sbjct: 808 LQSSGLTRGSVVDYPQPLATLHPYQSPPVRNFLGHNTSWLSQASIRGPWISSPTPAP--S 635
Query: 157 THLSASPSSDTIKLASVKGS-LPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTV 215
THLSASP DTIKL S KGS LPPSSSIK+VTPGPPASS+G Q VGT S LD NV V
Sbjct: 634 THLSASPVFDTIKLGSGKGSSLPPSSSIKNVTPGPPASSAGWQGILVGTASLLDVINVAV 455
Query: 216 PPAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKS 275
PAQ SS PK KKRKK V+SED GQK LQSLTPAV++ STS + TP+GNVP+++VEKS
Sbjct: 454 SPAQHSSDPKPKKRKKGVVSEDLGQKALQSLTPAVSNHTSTSFAVLTPLGNVPVTAVEKS 275
Query: 276 VVSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDK 335
+VSVSPL +QP+ND VEKRILSDESLMKVKEA+V+AEEASALS AAVNHSLELWNQLDK
Sbjct: 274 IVSVSPLDNQPENDGNVEKRILSDESLMKVKEAKVYAEEASALSGAAVNHSLELWNQLDK 95
Query: 336 HKNSGFMSDIEAKLASAAVAIAAAAAVAKAA 366
+KNS M D+EAKLASAAVA+AAAAA AKAA
Sbjct: 94 YKNSRSMPDVEAKLASAAVAVAAAAADAKAA 2
>CB828459
Length = 549
Score = 231 bits (588), Expect = 6e-61
Identities = 125/187 (66%), Positives = 143/187 (75%), Gaps = 11/187 (5%)
Frame = +2
Query: 784 KMSKNIDAAESANPDEMRSLNLTENEIVFNIGKSSTNESKQDPQRQVRSGLQKEGSKVIF 843
K SK+ D ESAN DE R LNLTENE VFNIGKSS NESKQDP R R+GLQKEGS+VIF
Sbjct: 2 KYSKSSDGVESANTDETRLLNLTENERVFNIGKSSKNESKQDPHRMARTGLQKEGSRVIF 181
Query: 844 GVPKPGKKRKFMEVSKHYVAHGSSKVNDKNDSVKIANFSMPQGSELRGWRNSSKNDSKEK 903
GVPKPGKKRKFM+VS+HYV HG + +ND++DSVKI N S GS + W+NSS+ND+KEK
Sbjct: 182 GVPKPGKKRKFMDVSQHYVEHGKNNINDRSDSVKIPNLS---GSLV--WKNSSRNDTKEK 346
Query: 904 LGADSKPKTKFGKPPGVLGRVNPPRNTSVSN-----------TEMNKDSSNHTKNASQSE 952
GA SKPKTKFGKP VLGRV PP++ S+SN TE DSSNH K+ASQSE
Sbjct: 347 HGA-SKPKTKFGKPESVLGRVIPPKDISLSNASSTTTDLTGQTERTMDSSNHLKSASQSE 523
Query: 953 SRVERAP 959
S+VERAP
Sbjct: 524 SQVERAP 544
>TC9049 similar to UP|O23575 (O23575) G2484-1 protein (Fragment), partial
(3%)
Length = 721
Score = 112 bits (281), Expect = 2e-25
Identities = 60/87 (68%), Positives = 66/87 (74%)
Frame = +1
Query: 984 PTKRTFTSRASKGKLAPASDKLRKGGGGKALNDKPTTSTSEPDALEPRRSNRRIQPTSRL 1043
PTK+ TSRASKGKLAPA DKL KG K +ND P+ TS D RRSNRRIQPTSRL
Sbjct: 13 PTKKVSTSRASKGKLAPAGDKLGKGEVEKTINDNPSKPTS--DVTAQRRSNRRIQPTSRL 186
Query: 1044 LEGLQSSLMVSKIPSVSHNRNIPKGEH 1070
LEGLQSSL++SKIPS SHNRN KG +
Sbjct: 187 LEGLQSSLIISKIPSFSHNRNTSKGNN 267
>AW428659
Length = 281
Score = 68.9 bits (167), Expect = 4e-12
Identities = 44/91 (48%), Positives = 54/91 (58%), Gaps = 9/91 (9%)
Frame = +2
Query: 919 GVLGRVNPPR---------NTSVSNTEMNKDSSNHTKNASQSESRVERAPYSTTDGATQV 969
G LGR PPR N S +E KDSS+H KNASQSE+++ GA
Sbjct: 14 GGLGRAIPPREKPPINSHTNDKTSRSERIKDSSSHFKNASQSENQMVG-----NTGAGAG 178
Query: 970 PIVFSSQATSTNTLPTKRTFTSRASKGKLAP 1000
P ++SS +ST++ PTKRT T RASKGKLAP
Sbjct: 179 PTLYSSLGSSTDSHPTKRTSTLRASKGKLAP 271
>TC7897 weakly similar to UP|ENL2_ARATH (Q9T076) Early nodulin-like protein
2 precursor (Phytocyanin-like protein), partial (31%)
Length = 1482
Score = 49.7 bits (117), Expect = 3e-06
Identities = 47/160 (29%), Positives = 66/160 (40%), Gaps = 4/160 (2%)
Frame = +3
Query: 62 SPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSALARGSVVDYSQALTP----L 117
SP S + PT P P +P LP+ + + + + G+ S + +P
Sbjct: 567 SPKASPPSGSPPTAGEPS-PAGTPPSVLPSPAGAPVPAGGPSAGTPPSASTSPSPSPVSA 743
Query: 118 HPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNTHLSASPSSDTIKLASVKGSL 177
P SP+P S+S S +P GP SP+PA D+ +PS T G
Sbjct: 744 PPAASPAP-GAESPSSSPTSNSPAGGPTATSPSPAGDSPAGGPPAPSPTT-------GDT 899
Query: 178 PPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPP 217
PPS+S GP S SG S+ +NN T PP
Sbjct: 900 PPSAS------GPDVSPSGTPSSLPADTPSSSSNNSTAPP 1001
>TC7812 UP|Q9LKJ6 (Q9LKJ6) Water-selective transport intrinsic membrane
protein 1, complete
Length = 1149
Score = 41.6 bits (96), Expect = 7e-04
Identities = 34/111 (30%), Positives = 46/111 (40%), Gaps = 5/111 (4%)
Frame = +1
Query: 119 PYQSPSPRNFLGHSTSWISQAPL---RGPWIGSPTPAPDNNTHLSASPSSDTIKLASVKG 175
P Q+P + +L S S +P P SPT AP S PS +S
Sbjct: 118 PTQTP*GQPWLSSSPPSSSSSPAPVPESPTTNSPTTAPPPPPASSPPPSPTHSPSSSPSP 297
Query: 176 SLPPSSSIKDVTPGPPASSSGL--QSTFVGTDSQLDANNVTVPPAQQSSGP 224
S P S + P P ASSS + S+ V + S L ++ + PP S P
Sbjct: 298 SAPTSPAATSTPPSPSASSSAVTSPSSAVSSTSSLSFSDPSSPPCSSPSSP 450
Score = 28.1 bits (61), Expect = 7.8
Identities = 32/132 (24%), Positives = 47/132 (35%), Gaps = 3/132 (2%)
Frame = +1
Query: 66 RASSKATPTIANPLIPLSSP---LWSLPTLSADSLQSSALARGSVVDYSQALTPLHPYQS 122
R S +P++ P +P W + + S + + + P P S
Sbjct: 73 RCRSATSPSVPLRRQPTQTP*GQPWLSSSPPSSSSSPAPVPESPTTNSPTTAPPPPPASS 252
Query: 123 PSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNTHLSASPSSDTIKLASVKGSLPPSSS 182
P P S+S AP P S P+P ++ SPSS +S+ S P S
Sbjct: 253 PPPSPTHSPSSSPSPSAPT-SPAATSTPPSPSASSSAVTSPSSAVSSTSSLSFSDPSS-- 423
Query: 183 IKDVTPGPPASS 194
PP SS
Sbjct: 424 -------PPCSS 438
>TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;}, partial (44%)
Length = 700
Score = 40.8 bits (94), Expect = 0.001
Identities = 37/143 (25%), Positives = 60/143 (41%)
Frame = -2
Query: 55 LQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSALARGSVVDYSQAL 114
L+G SSP SS P++++ + +P +S +LS S SS+ ++ S A
Sbjct: 447 LEGLSSSSPPSSESSSPEPSLSDLFLFDGTPAFSPSSLSLSSPSSSSGSKSLDASSSSAA 268
Query: 115 TPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNTHLSASPSSDTIKLASVK 174
+ + P++ S+S S + P P P + S+S SS +
Sbjct: 267 ATVRRTRYSVPKSSSSSSSSSSSSSSSSSSSGSPPPPRPPSTPPSSSSSSSSSSSSPPPG 88
Query: 175 GSLPPSSSIKDVTPGPPASSSGL 197
S SSS +P +SSS L
Sbjct: 87 ASSSSSSSPSSSSPSSSSSSSPL 19
Score = 30.0 bits (66), Expect = 2.1
Identities = 26/114 (22%), Positives = 48/114 (41%)
Frame = -2
Query: 150 TPAPDNNTHLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLD 209
TPA ++ +SPSS + + S +++++ P SSS S+ + S
Sbjct: 360 TPAFSPSSLSLSSPSSSSGSKSLDASSSSAAATVRRTRYSVPKSSSSSSSSSSSSSSSSS 181
Query: 210 ANNVTVPPAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATP 263
++ PP S+ P + S S P +S +S+S S+++P
Sbjct: 180 SSGSPPPPRPPSTPPSSSSSSSSSSS---------SPPPGASSSSSSSPSSSSP 46
>TC16422 similar to PIR|T05653|T05653 amino acid transport protein homolog
F22I13.20 - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (38%)
Length = 685
Score = 39.3 bits (90), Expect = 0.003
Identities = 49/183 (26%), Positives = 78/183 (41%), Gaps = 12/183 (6%)
Frame = +3
Query: 23 SVVKNLILIPQKHLCSQDLAARTSDSTVKQSVLQGKGISSP---LGRASSKATPTIANPL 79
S+ NLI+ Q+ C + + T L+ +SSP L +SK +PT ++P
Sbjct: 45 SIFSNLIISQQQQWCFRKNQKQVPPPTPSHPTLEKTHLSSPNPHLSPPNSKPSPTSSSPS 224
Query: 80 IPLSSPLWSLPTLSADSLQSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQA 139
S P SA SL +S+ GS P SPSP + +T+ S +
Sbjct: 225 --------SAPESSA-SLTASSEPAGS---------PASSCSSPSPSS---PTTA*CSSS 341
Query: 140 PLRG---PWIGSP------TPAPDNNTHLSASPSSDTIKLASVKGSLPPSSSIKDVTPGP 190
PL P +G+P T A + +ASPS+ + + + SSS + ++
Sbjct: 342 PLAASSIPSLGTPNSDPSATSASPSAARSAASPSTP*LSSPRLDSASATSSSSRPLSASS 521
Query: 191 PAS 193
PA+
Sbjct: 522 PAT 530
>TC11354 similar to UP|CAE54480 (CAE54480) Alpha glucosidase II , partial
(6%)
Length = 583
Score = 38.9 bits (89), Expect = 0.004
Identities = 42/135 (31%), Positives = 50/135 (36%), Gaps = 3/135 (2%)
Frame = -1
Query: 160 SASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQ 219
SA PSS K +S P S+S D P PP SS S S ++ T PP
Sbjct: 439 SAPPSSPGRKKSSETAIRPHSASEPDPAPPPPPPSSPPMSPSPTATSPPTSSPNTHPPTP 260
Query: 220 QS---SGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSV 276
S P S K L S P AS + TS S ++P P S S
Sbjct: 259 TPNP*SSPSPSTTTAS--SASPSTKTLPSTRPRPASESPTSSSPSSP---PPSSGSSVSP 95
Query: 277 VSVSPLADQPKNDQT 291
+SP P N T
Sbjct: 94 PRISPPPPPPSNSPT 50
Score = 37.4 bits (85), Expect = 0.013
Identities = 34/115 (29%), Positives = 48/115 (41%), Gaps = 4/115 (3%)
Frame = -1
Query: 116 PLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSP----TPAPDNNTHLSASPSSDTIKLA 171
P P SPSP +TS + +P P +P +P+P T SASPS+ T+
Sbjct: 340 PSSPPMSPSPT-----ATSPPTSSPNTHPPTPTPNP*SSPSPSTTTASSASPSTKTLPST 176
Query: 172 SVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQSSGPKA 226
+ P S S +P P SSG S + ++ PP S+ P A
Sbjct: 175 RPR---PASESPTSSSPSSPPPSSG---------SSVSPPRISPPPPPPSNSPTA 47
Score = 29.3 bits (64), Expect = 3.5
Identities = 29/100 (29%), Positives = 43/100 (43%), Gaps = 4/100 (4%)
Frame = -1
Query: 72 TPTIANPLIPLSSPLWSLPTLSADSLQSSALARGSVVDYSQALTPLHPYQSPSPRNFLGH 131
+PT +P P SSP PT + + S + + + S + L P P P +
Sbjct: 316 SPTATSP--PTSSPNTHPPTPTPNP*SSPSPSTTTASSASPSTKTL-PSTRPRPASESPT 146
Query: 132 STSWISQAPLRGPWIG----SPTPAPDNNTHLSASPSSDT 167
S+S S P G + SP P P +N+ + PSS T
Sbjct: 145 SSSPSSPPPSSGSSVSPPRISPPPPPPSNSPTATPPSSAT 26
>TC15031 weakly similar to UP|Q69107 (Q69107) LAT protein, partial (27%)
Length = 997
Score = 37.4 bits (85), Expect = 0.013
Identities = 29/112 (25%), Positives = 49/112 (42%), Gaps = 4/112 (3%)
Frame = +3
Query: 150 TPAPDNNTHLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLD 209
+P+ ++T +S +P+ I AS SSS + GP +SSG S+ GT
Sbjct: 525 SPSVSSSTFVS-NPNPSPISKASTASGPSVSSSSSSSSSGPSPNSSGYPSSAAGTSPPSP 701
Query: 210 ANNVTVPPAQQ----SSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTS 257
++ SS P++ R + ++ + P++ASRAS S
Sbjct: 702 IKPISAESTSS*IWASSNPRSSSRSSSSSTHRSRRRRRKKPLPSLASRASPS 857
>TC8484 similar to UP|Q9XGP1 (Q9XGP1) ESTs AU070372(S13446), partial (76%)
Length = 1121
Score = 37.0 bits (84), Expect = 0.017
Identities = 34/129 (26%), Positives = 51/129 (39%), Gaps = 6/129 (4%)
Frame = +1
Query: 131 HSTSWISQAPLRGPWIGSPTPA------PDNNTHLSASPSSDTIKLASVKGSLPPSSSIK 184
H+ W Q+P G ++ P+P P + + SPSS T+ + S PP
Sbjct: 82 HTL*WHLQSPYPGSFLPPPSPPLSSHPNPSSPPNSPPSPSSSTVATPNPSDSHPP----- 246
Query: 185 DVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQSSGPKAKKRKKDVLSEDHGQKLLQ 244
P PP S S A N PP S+ P A+ + L+ G++
Sbjct: 247 --PPSPPPSPS--------------ATNSLNPPYPTSTTP-ARSKPSPSLNSPRGRRQSS 375
Query: 245 SLTPAVASR 253
S +PA + R
Sbjct: 376 SPSPARSPR 402
>TC9679 weakly similar to UP|Q8H1B5 (Q8H1B5) Hin1-like protein, partial
(54%)
Length = 710
Score = 35.0 bits (79), Expect = 0.064
Identities = 28/124 (22%), Positives = 47/124 (37%)
Frame = +3
Query: 149 PTPAPDNNTHLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQL 208
P+ P T A ++D A+ S S+S PP+SSS S+ G+ S
Sbjct: 96 PSLRPKKPTTAQAEAAADASSAAAAASSASSSNS-----SSPPSSSSASPSSSSGSSSAP 260
Query: 209 DANNVTVPPAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVP 268
+ + T P + S + + +S Q+ + S V +G +P
Sbjct: 261 TS*SSTSPTPRSRSSTSPETTRSTTISPSTSPSATQTSASGSTTTTSKRVRCTKTLGLIP 440
Query: 269 MSSV 272
SS+
Sbjct: 441 NSSI 452
>TC16353 weakly similar to UP|HQGT_ARATH (Q9M156) Probable hydroquinone
glucosyltransferase (Arbutin synthase) , partial (52%)
Length = 885
Score = 34.7 bits (78), Expect = 0.084
Identities = 39/154 (25%), Positives = 64/154 (41%)
Frame = +2
Query: 71 ATPTIANPLIPLSSPLWSLPTLSADSLQSSALARGSVVDYSQALTPLHPYQSPSPRNFLG 130
++P+ ++ SP S P L L+S + R + ++ + P S SP +
Sbjct: 122 SSPSDSSSTTTSKSPSSSPPKLHRQKLKSPSSNRFRIPFHTFSFLP-----STSPTSRKT 286
Query: 131 HSTSWISQAPLRGPWIGSPTPAPDNNTHLSASPSSDTIKLASVKGSLPPSSSIKDVTPGP 190
+ S +P P+ S TP+ + S+S SS S + S PP +S P P
Sbjct: 287 P*LKYESPSPYSAPFRPSATPSAPSPPPHSSSTSSALTPSTSPENSAPPLTS--SSLPPP 460
Query: 191 PASSSGLQSTFVGTDSQLDANNVTVPPAQQSSGP 224
P S S ST + + A++ P +S P
Sbjct: 461 PRSLS--SSTSLAWTGRFIASSENSPNRSKSPAP 556
>BP033767
Length = 541
Score = 34.7 bits (78), Expect = 0.084
Identities = 22/58 (37%), Positives = 26/58 (43%), Gaps = 9/58 (15%)
Frame = +1
Query: 178 PPSSSIKDVTPGPPASSSG---------LQSTFVGTDSQLDANNVTVPPAQQSSGPKA 226
PPSSS+K P PP SSS S+ S DA T PP ++ S P A
Sbjct: 364 PPSSSLKPPNPNPPTSSSASPLPPSPAPSPSSLSPPSSTADAAAKTTPPTRRLSDPTA 537
>TC7994 similar to UP|Q8RU73 (Q8RU73) Ribose-5-phosphate isomerase
precursor , partial (81%)
Length = 1355
Score = 34.7 bits (78), Expect = 0.084
Identities = 37/154 (24%), Positives = 58/154 (37%), Gaps = 5/154 (3%)
Frame = +2
Query: 116 PLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTP---APDNNTHLSASPSSDTIKLAS 172
P Y P P N+ + PL P + SP+P + D+ ++ SS AS
Sbjct: 317 PTLSYAPPLPLNY--------APPPLPNPSVPSPSPKTTSRDSPPTRPSTTSSPAWSSAS 472
Query: 173 VKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQSSGPKAKKRKKD 232
PPSSS + PPA+S P + S P A++ ++D
Sbjct: 473 APAPPPPSSSPSSASFSPPANS---------------------PTSSASPPPSARRSRRD 589
Query: 233 --VLSEDHGQKLLQSLTPAVASRASTSVSAATPV 264
+ S++P+ A ST S ++ V
Sbjct: 590 PSASRSPSSTTIPASISPSTAPTRSTLSSTSSRV 691
>TC9392
Length = 650
Score = 34.7 bits (78), Expect = 0.084
Identities = 17/60 (28%), Positives = 32/60 (53%)
Frame = +1
Query: 687 WSVGDRVDAWIQESWREGVITEKNKKDETTLTVHIPASGETSVLRAWNLRPSLIWKDGQW 746
+ +GD+VDA+ + SWREG + ++ + + + ++P + + NLR W D W
Sbjct: 250 FKIGDKVDAYDKGSWREGHLVKELEDGKFAVDFNLPK--QLNEFPKENLRTHREWIDDHW 423
Score = 31.2 bits (69), Expect = 0.92
Identities = 14/40 (35%), Positives = 23/40 (57%), Gaps = 1/40 (2%)
Frame = +1
Query: 594 EENTFKEGSLVEVFKDEEGHKAAWFMGNILSLK-DGKVYV 632
++ F+ G LVEV +G++ WF+ ++ LK GK V
Sbjct: 445 KQELFRIGDLVEVSSKVKGYRGTWFLAEVVELKVQGKFLV 564
>TC11455 similar to UP|Q9ZUS8 (Q9ZUS8) At2g37380 protein, partial (10%)
Length = 575
Score = 33.9 bits (76), Expect = 0.14
Identities = 40/157 (25%), Positives = 54/157 (33%), Gaps = 3/157 (1%)
Frame = +2
Query: 75 IANPLIPLSSPLWSLPTLSADSLQSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTS 134
+ NP + L PL L +++S S LA P S P N ++S
Sbjct: 101 LQNPTLSLQHPLTLLHLPTSNSPSPSHLAN-------------LPPHSALPMNSSTKASS 241
Query: 135 WISQAPLRGPWIG---SPTPAPDNNTHLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPP 191
++S PW SP P P H P + A + PSS TP P
Sbjct: 242 YLSTFLPASPWSAPSSSPPPPPHPPPHPPPPPLETPLAAAHPSTATSPSS--PTATPPAP 415
Query: 192 ASSSGLQSTFVGTDSQLDANNVTVPPAQQSSGPKAKK 228
A S T S D + T S P ++K
Sbjct: 416 APSQ-------RTTSSSDFTSTTATTTTHYSNPTSRK 505
>TC10758
Length = 532
Score = 33.9 bits (76), Expect = 0.14
Identities = 29/102 (28%), Positives = 40/102 (38%), Gaps = 16/102 (15%)
Frame = +2
Query: 115 TPLHPYQSPSPRNFLGHST-------SWISQAP-------LRGPWIGSPTPAPDNNTHLS 160
TP HP SPSP+ S+ SW P P SPT P + S
Sbjct: 143 TPSHPQPSPSPQKISARSSNPSPRTNSWSFSNPPPSVTPTYSTPSAPSPTATPPSENSSS 322
Query: 161 ASPSSDTIKLASVKGSLPPSSSIKDVTP--GPPASSSGLQST 200
A+ + S SLP ++S K ++ PPA + S+
Sbjct: 323 AASPAKPPPRRSAASSLPTATSTKRLSSSINPPAGPRVMASS 448
>BP039723
Length = 587
Score = 33.9 bits (76), Expect = 0.14
Identities = 35/109 (32%), Positives = 47/109 (43%), Gaps = 9/109 (8%)
Frame = -2
Query: 136 ISQAPLRGPWIGSPTPAPDNNTHLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSS 195
IS+ P GP IGSPTP +L SP S A P SS TP P ++ S
Sbjct: 484 ISEPPSPGPAIGSPTP------NLPPSPESG---FAPASQPSPAPSSGPSSTPSPASAPS 332
Query: 196 GL-------QSTFVGTDSQLDANNVTVPPAQQSSGPKAK--KRKKDVLS 235
S+F + A +V+ PP+ S+ + K RK VL+
Sbjct: 331 PKPLVPHLPPSSFFPKLTPPAAEDVSAPPSSDSNKQEDKHNNRKTVVLA 185
>TC13802 similar to UP|Q9M1T1 (Q9M1T1) Sugar-phosphate isomerase-like
protein, partial (41%)
Length = 522
Score = 33.9 bits (76), Expect = 0.14
Identities = 36/146 (24%), Positives = 55/146 (37%), Gaps = 4/146 (2%)
Frame = +2
Query: 83 SSPLWSLPTLSADSLQSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLR 142
+ PL S T + S+ S + TP P SP+P + L +AP
Sbjct: 53 NKPLHSTLTKPPSQISSNPNKTTSTTSSPTSTTP-KPSPSPAPSSTL--------RAPSS 205
Query: 143 GPWIGSPTPAPDNNTHLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQ---- 198
P SP +P S PSS + L PS++ + +P +SSS
Sbjct: 206 SPASASPASSPTK----SPKPSSPSASAPPSSTPLTPSTATSESSPTATSSSSSASPAPP 373
Query: 199 STFVGTDSQLDANNVTVPPAQQSSGP 224
++F L+ + P+ SS P
Sbjct: 374 TSFSALSRALELKALASSPSPPSSPP 451
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.308 0.125 0.347
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,881,174
Number of Sequences: 28460
Number of extensions: 241239
Number of successful extensions: 1686
Number of sequences better than 10.0: 139
Number of HSP's better than 10.0 without gapping: 1508
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1629
length of query: 1091
length of database: 4,897,600
effective HSP length: 100
effective length of query: 991
effective length of database: 2,051,600
effective search space: 2033135600
effective search space used: 2033135600
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC137077.8


