

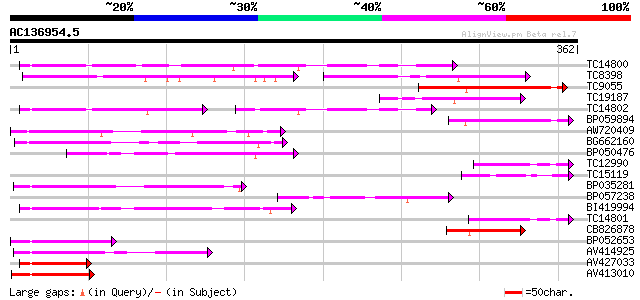
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136954.5 - phase: 0
(362 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC14800 weakly similar to UP|Q9FHP3 (Q9FHP3) Genomic DNA, chromo... 114 3e-26
TC8398 weakly similar to UP|Q84KK1 (Q84KK1) S haplotype-specific... 70 2e-22
TC9055 82 2e-16
TC19187 69 9e-13
TC14802 weakly similar to UP|CAD56661 (CAD56661) S locus F-box (... 65 2e-11
BP059894 62 1e-10
AW720409 62 2e-10
BG662160 61 3e-10
BP050476 59 2e-09
TC12990 56 8e-09
TC15119 55 1e-08
BP035281 55 2e-08
BP057238 54 3e-08
BI419994 53 9e-08
TC14801 50 8e-07
CB826878 49 1e-06
BP052653 49 2e-06
AV414925 47 4e-06
AV427033 46 9e-06
AV413010 44 3e-05
>TC14800 weakly similar to UP|Q9FHP3 (Q9FHP3) Genomic DNA, chromosome 5, TAC
clone:K14B20, partial (7%)
Length = 782
Score = 114 bits (285), Expect = 3e-26
Identities = 99/286 (34%), Positives = 139/286 (47%), Gaps = 6/286 (2%)
Frame = +3
Query: 7 LPVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLSKSKGNLRLALFSNK 66
LP ELI EIL V SL+ + VSK W + ISDP FVK+HL + + R A F +
Sbjct: 48 LPDELII-EILSWLPVKSLLQFRVVSKTWKSFISDPQFVKLHL---LHRLSFRNADFEHT 215
Query: 67 NLRLQIRAGGRGCSYTVTVAPTSVSLLLESTTSSIPIADDLQYQFSCVDCCGIIGSCNGL 126
+L ++ G Y ++ +VS LLES ++ + SC+ IG+CNGL
Sbjct: 216 SLLIKCHTDDFGRPY---ISSRTVSSLLESPSAIVA-------SRSCISGYDFIGTCNGL 365
Query: 127 ICLHGCFHGSGYKKH--SFCFWNPATRSKSKTLLYVPSYLNR-VRLGFGYDNSTDTYKTV 183
+ L + + FWNPATR+ S+ PS+ R + LGFGYD S+DTYK V
Sbjct: 366 VSLRKLNYDESNTNNFSQVRFWNPATRTMSQD--SPPSWSPRNLHLGFGYDCSSDTYKVV 539
Query: 184 --MFGITMDEGLGGNRMRTAVVKVFTLGDSIWRDIQSSFPVELALRSRWDDIKYDGVYLS 241
+ G+TM V V+ +GD+ WR IQ S + L+ VY+S
Sbjct: 540 GMIPGLTM-------------VNVYNMGDNCWRTIQISPHAPMHLQG-------SAVYVS 659
Query: 242 NSISWLVCHRYKCQQKNLTTEQFVIISLDLETETYT-QLQLPKLPF 286
N+++WL T + I+S DLE E QL LP P+
Sbjct: 660 NTLNWLA-----------GTNPYFIVSFDLEKEECAQQLSLPYCPW 764
>TC8398 weakly similar to UP|Q84KK1 (Q84KK1) S haplotype-specific F-box
protein a, partial (9%)
Length = 1602
Score = 70.1 bits (170), Expect(2) = 2e-22
Identities = 45/140 (32%), Positives = 70/140 (49%), Gaps = 8/140 (5%)
Frame = +1
Query: 201 AVVKVFTLGDSIWRDIQSSFPVELALRSRWDDIKYDGVYLSNSISWLVCHRYKCQQKNLT 260
+V V+ +GD WR IQS R +GVYL+ +I+W++ + N+
Sbjct: 727 SVANVYNMGDGCWRRIQSFPDFTREHWGRGTAPDQNGVYLNGTINWVL-------RLNIA 885
Query: 261 TEQFVIISLDLETETYTQLQLPKLP-------FDNPNICALM-DCICFSYDFKETHFVIW 312
+ IISLDL +E ++L LP P +D P I ++ D +C ++ + FVIW
Sbjct: 886 YD---IISLDLGSEVCSRLSLPCFPPPGPKCVYDLPPILGVLKDSLCVLHNNNDKTFVIW 1056
Query: 313 QMKEFGVEESWTQFLKISYQ 332
+M EFG ESWT ++
Sbjct: 1057KMNEFGAHESWTPLFNFDFK 1116
Score = 52.0 bits (123), Expect(2) = 2e-22
Identities = 60/205 (29%), Positives = 88/205 (42%), Gaps = 29/205 (14%)
Frame = +3
Query: 9 VELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLSKSKGNLRLALFSNKNL 68
++ + EIL V SL+ + V K W IS+P F+K+HL+ S +K A F++ +
Sbjct: 81 IDELIWEILSLLPVKSLVRFRCVCKSWKLTISNPQFMKLHLRRSSAKA---AADFADSQV 251
Query: 69 RLQIRAGGRGCSYTVTV------------APTSVSLLLESTTS---SIPIADDL---QYQ 110
+ + S ++ P V L E SI A L Q
Sbjct: 252 VVMTKRDHIAHSPLLSKFHIVHDETFTADEPWPVHALREPFNQPHVSICYAPSLFSSQNN 431
Query: 111 FSCVDCCGIIGSCNGLICL--HGCFHGSGYKKHSFCFWNPATRSKSK-----TLLYVP-- 161
C +IG+CNGL+ + G K S C NPAT+ +S+ +L Y
Sbjct: 432 DKLKGHCRLIGACNGLVSVINEGYNSAESATKLSCCNLNPATKLRSQVSPTLSLCYKQFH 611
Query: 162 SYLNRVR--LGFGYDNSTDTYKTVM 184
S ++ V GFGYD DTYK V+
Sbjct: 612 SLIHSVFRIFGFGYDPLNDTYKVVV 686
>TC9055
Length = 577
Score = 81.6 bits (200), Expect = 2e-16
Identities = 43/97 (44%), Positives = 60/97 (61%), Gaps = 2/97 (2%)
Frame = +2
Query: 262 EQFVIISLDLETETYTQLQLPKLPFDNPN--ICALMDCICFSYDFKETHFVIWQMKEFGV 319
EQ VI+SLD+ E Y L LP+ + P+ I L +C+C YDFK THFV W+M E+G+
Sbjct: 41 EQLVIVSLDMRQEAYRLLSLPQGTSELPSAEIQVLGNCLCLFYDFKRTHFVAWKMSEYGL 220
Query: 320 EESWTQFLKISYQNLGINYILGRNGFLVLPVCLSENG 356
ESWT L ISY++L ++ N +L +CL +G
Sbjct: 221 PESWTPLLTISYEHLRWDHRFYLNQWL---ICLCGDG 322
>TC19187
Length = 632
Score = 69.3 bits (168), Expect = 9e-13
Identities = 34/96 (35%), Positives = 54/96 (55%), Gaps = 3/96 (3%)
Frame = +1
Query: 237 GVYLSNSISWLVCHRYKCQQKNLTTEQFVIISLDLETETYTQLQLP---KLPFDNPNICA 293
G ++S +++W Y +L ++I+SLDL+ ETY ++ P K P +
Sbjct: 7 GKFVSGTLNWAA--NYSVGVSSL----WIIVSLDLQKETYKEILPPDYEKEECSTPTLSV 168
Query: 294 LMDCICFSYDFKETHFVIWQMKEFGVEESWTQFLKI 329
L C+C +YD K T FV+W MK++GV ESW + + I
Sbjct: 169 LKGCLCMNYDHKRTDFVVWMMKDYGVRESWVKLVTI 276
>TC14802 weakly similar to UP|CAD56661 (CAD56661) S locus F-box (SLF)-S4
protein, partial (10%)
Length = 551
Score = 64.7 bits (156), Expect = 2e-11
Identities = 48/131 (36%), Positives = 66/131 (49%), Gaps = 3/131 (2%)
Frame = +3
Query: 145 FWNPATRSKSKTLLYVPSYLNR-VRLGFGYDNSTDTYKTV--MFGITMDEGLGGNRMRTA 201
FWNPATR+ S+ PS+ R + LGFGYD S+DTYK V + G+TM
Sbjct: 255 FWNPATRTMSQDS--PPSWSPRNLHLGFGYDCSSDTYKVVGMIPGLTM------------ 392
Query: 202 VVKVFTLGDSIWRDIQSSFPVELALRSRWDDIKYDGVYLSNSISWLVCHRYKCQQKNLTT 261
V V+ +GD+ WR IQ S + L+ VY+SN+++WL T
Sbjct: 393 -VNVYNMGDNCWRTIQISPHAPMHLQG-------SAVYVSNTLNWLA-----------GT 515
Query: 262 EQFVIISLDLE 272
+ I+S DLE
Sbjct: 516 NPYFIVSFDLE 548
Score = 42.4 bits (98), Expect = 1e-04
Identities = 40/126 (31%), Positives = 59/126 (46%), Gaps = 6/126 (4%)
Frame = +3
Query: 7 LPVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLSKSKGNLRLALFSNK 66
LP ELI EIL V SL+ + VSK W + ISDP FVK+HL + + R A F +
Sbjct: 21 LPDELII-EILSWLPVKSLLQFRVVSKTWKSFISDPQFVKLHL---LHRLSFRNADFEHT 188
Query: 67 NLRLQIRAGGRGCSYTVTVA-----PTSVSLLLESTTSSIPIADDLQYQFSC-VDCCGII 120
+L ++ G Y + P + ++ +S S P L + + C D ++
Sbjct: 189 SLLIKCHTDDFGRPYISSRTVRFWNPATRTMSQDSPPSWSPRNLHLGFGYDCSSDTYKVV 368
Query: 121 GSCNGL 126
G GL
Sbjct: 369 GMIPGL 386
>BP059894
Length = 474
Score = 62.4 bits (150), Expect = 1e-10
Identities = 35/82 (42%), Positives = 49/82 (59%), Gaps = 2/82 (2%)
Frame = -3
Query: 281 LPKLPFDNP--NICALMDCICFSYDFKETHFVIWQMKEFGVEESWTQFLKISYQNLGINY 338
LPK + P I L +C+C +D THFV W+M E+GV ESWT+ L ISYQ+ N
Sbjct: 469 LPKGTSELPYVEIQVLGNCLCLFHDDNRTHFVGWKMTEYGVPESWTRMLSISYQHFQCND 290
Query: 339 ILGRNGFLVLPVCLSENGATLI 360
++ R+ +L VCL +G L+
Sbjct: 289 VILRHRWL---VCLCGDGNILM 233
>AW720409
Length = 575
Score = 62.0 bits (149), Expect = 2e-10
Identities = 67/191 (35%), Positives = 84/191 (43%), Gaps = 15/191 (7%)
Frame = +3
Query: 1 MNISEILPVELITTEILLRPDVNSLMLLKFVSKPWNTLI-SDPIFVKMHLKLSKSKGN-- 57
M I +LP EL EIL V +LM VSK W +LI D F K+HL S N
Sbjct: 75 MEIEFVLPFEL-WIEILSWLPVKTLMRFSCVSKSWKSLIYQDRDFKKLHLDRSPKNNNQV 251
Query: 58 ---LRLALFSNKNLRLQIRAGGRGCSYTVTVAPTSVSLLLESTT-SSIPIADDLQYQFSC 113
L+ LFS + T P V LL+ SS I DD+ ++
Sbjct: 252 ILTLQKPLFSGR-----------------TFFPFPVRRLLQDQEPSSSSIIDDIPFEEED 380
Query: 114 VD----CCGIIGSCNGLICLHGC-FHGSGYKKHSFCFWNPATR---SKSKTLLYVPSYLN 165
D IGSCNGL+CL+G HG ++ WNPATR KS L V +
Sbjct: 381 EDEEDNYHDTIGSCNGLVCLYGINDHGLWFR-----LWNPATRFRFHKSPPLDAVIG--S 539
Query: 166 RVRLGFGYDNS 176
+ FGYD+S
Sbjct: 540 TLHFAFGYDHS 572
>BG662160
Length = 445
Score = 61.2 bits (147), Expect = 3e-10
Identities = 57/178 (32%), Positives = 78/178 (43%), Gaps = 4/178 (2%)
Frame = +2
Query: 4 SEILPVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLSKSKG-NLRLAL 62
++ LP EL EIL V SL+ + VSK W ++ISD F+K+HL S S N A
Sbjct: 14 AQFLPEEL-RLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTTRNTDFAY 190
Query: 63 FSNKNLRLQIRAGGRGCSYTVTVAPTSVSLLLESTTSSIPIADDLQYQFSCVDCCGIIGS 122
+ + +P + S+I I DD + G+
Sbjct: 191 LQS-----------------LITSPRK-----SRSASTIAIPDDYDF----------CGT 274
Query: 123 CNGLICLHGCFHGSGYKKHSFC-FWNPATRSKSKTL--LYVPSYLNRVRLGFGYDNST 177
CNGL+CLH + K S WNPA RS S+ LY+P + GF YD+ST
Sbjct: 275 CNGLVCLHSSKYDRKDKYTSHVRLWNPAMRSMSQRSPPLYLPIVYDSY-FGFDYDSST 445
>BP050476
Length = 520
Score = 58.5 bits (140), Expect = 2e-09
Identities = 50/156 (32%), Positives = 69/156 (44%), Gaps = 8/156 (5%)
Frame = +2
Query: 37 TLISDPIFVKMHLKLSKSKGNLRLALFSNKNLRLQIRAGGRGCSYTVTVAPTSVSLLLES 96
T D FVK+HL S ++ L + +N + + V P+SV L+E
Sbjct: 5 TYHDDSSFVKLHLNRSPKNTHILLNI-ANDPYDFE--------NDDTWVVPSSVCCLIED 157
Query: 97 TTSSIPIADDLQYQFSCVDCCGIIGSCNGLICLHGCFHGSGYKKHSFCFWNPATRSKSK- 155
+S I D + + D +IGS NGLIC + ++ WNPAT KSK
Sbjct: 158 LSSMI----DAKGCYLLKDGHLVIGSSNGLICFGNFYDVGPIEEFWVQLWNPATHLKSKK 325
Query: 156 -------TLLYVPSYLNRVRLGFGYDNSTDTYKTVM 184
V + +V LGFGYDN DTYK V+
Sbjct: 326 SPTFNLSMRTSVDAPPGKVNLGFGYDNLHDTYKVVL 433
>TC12990
Length = 518
Score = 56.2 bits (134), Expect = 8e-09
Identities = 32/65 (49%), Positives = 38/65 (58%), Gaps = 1/65 (1%)
Frame = +1
Query: 297 CICFSYDFKETH-FVIWQMKEFGVEESWTQFLKISYQNLGINYILGRNGFLVLPVCLSEN 355
C+C S + KETH FV+WQMKEFGV ESWTQ I IL F +C+SEN
Sbjct: 16 CLCISQNNKETHSFVVWQMKEFGVHESWTQLFNIIVH----ERILHTPCF---AMCMSEN 174
Query: 356 GATLI 360
G L+
Sbjct: 175 GDALL 189
>TC15119
Length = 520
Score = 55.5 bits (132), Expect = 1e-08
Identities = 28/72 (38%), Positives = 43/72 (58%)
Frame = +3
Query: 289 PNICALMDCICFSYDFKETHFVIWQMKEFGVEESWTQFLKISYQNLGINYILGRNGFLVL 348
P + L +C+C ++F T +WQM++FGV ESWTQ LKI+ N+ +N +
Sbjct: 42 PTLVILKNCLCACHEFLVT---VWQMEKFGVRESWTQLLKIT--NIHLN---------AV 179
Query: 349 PVCLSENGATLI 360
P+C+SEN L+
Sbjct: 180 PLCMSENDVVLL 215
>BP035281
Length = 498
Score = 54.7 bits (130), Expect = 2e-08
Identities = 48/152 (31%), Positives = 70/152 (45%), Gaps = 3/152 (1%)
Frame = +2
Query: 3 ISEILPVELITTEILLRPDVNSLMLLKFVSKPWNTLIS-DPIFVKMHLKLSKSKGNLRLA 61
++++ P EL+ EIL V SL+ K V K W +LIS + F K+ L+ + K N +
Sbjct: 110 VTDVFPSELVI-EILSWLPVLSLIRFKCVCKSWKSLISHNKAFAKLQLERTSLKINHVIL 286
Query: 62 LFSNKNLRLQIRAGGRGCSYTVTVAPTSVSLLLESTTSSIPIADDLQYQFSCVDCCGIIG 121
S+ + P S+ LLE +S + D + + +IG
Sbjct: 287 TSSDSSF-----------------IPCSLPRLLEDPSSIMDQDQDRCLRLDKFEYNEVIG 415
Query: 122 SCNGLICLHGCFHGSGYKKHSFCF--WNPATR 151
SCNGLICLH + G FCF NP+TR
Sbjct: 416 SCNGLICLHADYCAPG-----FCFRPCNPSTR 496
>BP057238
Length = 434
Score = 54.3 bits (129), Expect = 3e-08
Identities = 37/115 (32%), Positives = 56/115 (48%), Gaps = 3/115 (2%)
Frame = -2
Query: 172 GYDNSTDTYKTVMFGITMDEGLGGNRMRTAVVKVFTLGDSIWRDIQSSFPVELALRSRWD 231
GYD S DTYK VM E +M T V+ +GDS WR I SS + L+
Sbjct: 433 GYDESRDTYKAVMALCHSIE--PEPKMETT---VYCMGDSCWRKISSSPSSSVLLQ---- 281
Query: 232 DIKYDGVYLSNSISWLVCHRY---KCQQKNLTTEQFVIISLDLETETYTQLQLPK 283
++DG ++ ++WL + +++T +Q VI+ + E YT L LP+
Sbjct: 280 --QFDGQFVGGCVNWLALDNLNGPNYEWQSVTLDQLVIVCFHMREEAYTYLSLPE 122
>BI419994
Length = 549
Score = 52.8 bits (125), Expect = 9e-08
Identities = 55/180 (30%), Positives = 74/180 (40%), Gaps = 3/180 (1%)
Frame = +2
Query: 7 LPVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLSKSKGNLRLALFSNK 66
LP ELI +ILLR V SL+ K V K W LISDP F K H L+ S + R+ L +
Sbjct: 47 LPDELIL-QILLRLPVRSLLRFKTVCKSWRYLISDPQFTKSHFNLAASPTH-RIFLNNTN 220
Query: 67 NLRLQIRAGGRGCSYTVTVAPTSVSLLLESTTSSIPIADDLQYQFSCVDCCGIIGSCNGL 126
++ S + S L+ P D S V ++GSC G
Sbjct: 221 GF*IE--------SLDTDSSLHDESSLVHLKFPLAPPHPDNHRFGSSVTGLKVLGSCRGF 376
Query: 127 ICLHGCFHGSGYKKHSFCFWNPATRSKSKTLLYVPSYLN---RVRLGFGYDNSTDTYKTV 183
+ L + + WNP+T + + + SYL G GYD S D Y V
Sbjct: 377 VLL-------ANHQGNVVVWNPSTGVRRRVPEWC-SYLASQCEFLYGIGYDESNDDYLVV 532
>TC14801
Length = 595
Score = 49.7 bits (117), Expect = 8e-07
Identities = 31/68 (45%), Positives = 37/68 (53%), Gaps = 1/68 (1%)
Frame = +2
Query: 294 LMDCICFSYDFKETH-FVIWQMKEFGVEESWTQFLKISYQNLGINYILGRNGFLVLPVCL 352
L C+C S KETH FV+ QMKE+GV ESWTQ I IL F +C+
Sbjct: 110 LRGCLCISQYHKETHSFVVSQMKEYGVNESWTQLFNIIVH----ERILHSPCF---AMCM 268
Query: 353 SENGATLI 360
SENG L+
Sbjct: 269 SENGDVLL 292
>CB826878
Length = 635
Score = 48.9 bits (115), Expect = 1e-06
Identities = 25/56 (44%), Positives = 36/56 (63%), Gaps = 6/56 (10%)
Frame = +3
Query: 280 QLPKLPFDNPNIC-----ALMDCICFSYDFKET-HFVIWQMKEFGVEESWTQFLKI 329
QLP+ ++P++ L D +C S + K+T +FV+WQMKEFGV +SWTQ I
Sbjct: 342 QLPEQCLNSPHLYMPILGVLRDSLCVSQNDKKTRNFVVWQMKEFGVHKSWTQLFNI 509
>BP052653
Length = 552
Score = 48.5 bits (114), Expect = 2e-06
Identities = 30/68 (44%), Positives = 40/68 (58%)
Frame = +1
Query: 1 MNISEILPVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLSKSKGNLRL 60
+N+S ILP +LI +ILLR V SL+ K V K W +LISDP F K H L+ + + L
Sbjct: 169 LNLSSILPHDLIV-QILLRLPVRSLLRFKSVCKSWLSLISDPQFGKSHFDLAAAPTHRCL 345
Query: 61 ALFSNKNL 68
+N L
Sbjct: 346 VKINNFKL 369
>AV414925
Length = 414
Score = 47.4 bits (111), Expect = 4e-06
Identities = 39/128 (30%), Positives = 60/128 (46%), Gaps = 1/128 (0%)
Frame = +1
Query: 3 ISEILPVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLSKSKGNLRLAL 62
++E +PVE+I +IL R V SL+ + +SK W +LI F+K+HL S + N L
Sbjct: 85 MAEEIPVEVIA-DILSRLPVTSLLRFRSISKSWRSLIDSKHFMKLHLNNSLTNPNPNLT- 258
Query: 63 FSNKNLRLQIRAGGRGCSYTVTVAPTSVSLLLESTTSSIPIADDLQYQFSCV-DCCGIIG 121
NL L+ L + SI A +L + C + I+G
Sbjct: 259 ----NLILRHNTD-----------------LYRADFPSIGAAVNLNHPLMCYSNRINILG 375
Query: 122 SCNGLICL 129
SC+GL+C+
Sbjct: 376 SCHGLLCI 399
>AV427033
Length = 420
Score = 46.2 bits (108), Expect = 9e-06
Identities = 25/46 (54%), Positives = 30/46 (64%)
Frame = +1
Query: 7 LPVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLS 52
LP +L+ EIL R VNSL+ + V K WN+LISDP F K HL S
Sbjct: 124 LPFDLVV-EILCRLPVNSLLQFRCVCKSWNSLISDPKFAKNHLHCS 258
>AV413010
Length = 416
Score = 44.3 bits (103), Expect = 3e-05
Identities = 26/53 (49%), Positives = 34/53 (64%)
Frame = +3
Query: 2 NISEILPVELITTEILLRPDVNSLMLLKFVSKPWNTLISDPIFVKMHLKLSKS 54
++S ILP ELI +ILL+ V SL+ K VSK W + IS+P F K H L+ S
Sbjct: 81 SLSSILPDELII-QILLQLPVTSLLRFKSVSKSWFSQISNPQFAKSHFNLAAS 236
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.324 0.140 0.436
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,581,709
Number of Sequences: 28460
Number of extensions: 126594
Number of successful extensions: 731
Number of sequences better than 10.0: 74
Number of HSP's better than 10.0 without gapping: 702
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 710
length of query: 362
length of database: 4,897,600
effective HSP length: 91
effective length of query: 271
effective length of database: 2,307,740
effective search space: 625397540
effective search space used: 625397540
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 56 (26.2 bits)
Medicago: description of AC136954.5


