

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
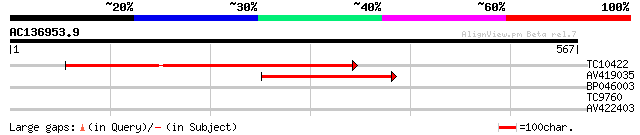
Query= AC136953.9 - phase: 0
(567 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC10422 similar to UP|Q944P7 (Q944P7) AT4g30920/F6I18_170, parti... 453 e-128
AV419035 250 4e-67
BP046003 33 0.095
TC9760 29 1.8
AV422403 27 6.8
>TC10422 similar to UP|Q944P7 (Q944P7) AT4g30920/F6I18_170, partial (47%)
Length = 874
Score = 453 bits (1166), Expect = e-128
Identities = 237/292 (81%), Positives = 260/292 (88%)
Frame = +3
Query: 56 PTNIEAPKISFSATDVDVTEWKGDILAVGVTEKDLTRDAKSRFENSILNKIDLKLDGLLS 115
P N+E KISF+ DVDVTEWKGDILAVGV EKDL RD K RFEN ILNKID KL GLL+
Sbjct: 6 PANVETHKISFTPKDVDVTEWKGDILAVGVAEKDLVRDGKLRFENVILNKIDSKLGGLLA 185
Query: 116 EASSEEDFSGKVGQSTVVRIKGLGSKRVGLIGLGQLPSTTALYKGLGEAVVAVAKSAQAS 175
EAS+EEDFSGK GQSTV+R+ GLG+KRV LIGLG S+TA +KGLGEAVVA AKS+QAS
Sbjct: 186 EASAEEDFSGKTGQSTVLRVTGLGAKRVSLIGLG---SSTAPFKGLGEAVVAAAKSSQAS 356
Query: 176 NVAIVLASSEGLSSESKLSTAYAIASGAVLGLFEDQRYKSESKKPAVRSIDIIGLGTGPD 235
+VA+VLASS+GLS+ESKLS+AYAIASG +LG FED RYKSESKK A+RS+DIIGLGTGP+
Sbjct: 357 SVAVVLASSDGLSAESKLSSAYAIASGLMLGFFEDNRYKSESKKLALRSVDIIGLGTGPE 536
Query: 236 LEKKLKYAGDVSFGIIFGRELVNSPANVLTPGVLAEEASKVASTYSDVFTAKILDADQCK 295
LEKKLKYAG VS GIIFGRELVNSPANVLTPG LAEEASKVAS+YSDVFTA IL+A+QCK
Sbjct: 537 LEKKLKYAGYVSSGIIFGRELVNSPANVLTPGGLAEEASKVASSYSDVFTATILNAEQCK 716
Query: 296 ELKMGSYLGVAAASANPPRFIHLTYKPPSGSVKVKLALVGKGLTFDSGGYNI 347
ELKMGSYLGVAAASANPP FIHL YKPP+G V VKLALVGKGLTFDSGGYNI
Sbjct: 717 ELKMGSYLGVAAASANPPHFIHLRYKPPTGPVNVKLALVGKGLTFDSGGYNI 872
>AV419035
Length = 407
Score = 250 bits (639), Expect = 4e-67
Identities = 125/135 (92%), Positives = 127/135 (93%)
Frame = +1
Query: 252 FGRELVNSPANVLTPGVLAEEASKVASTYSDVFTAKILDADQCKELKMGSYLGVAAASAN 311
FGRELVNSPANVLTPGVLAEEASK+ASTYSDVFTAKILDA+QCKELKMGSYL VAAASAN
Sbjct: 1 FGRELVNSPANVLTPGVLAEEASKIASTYSDVFTAKILDAEQCKELKMGSYLAVAAASAN 180
Query: 312 PPRFIHLTYKPPSGSVKVKLALVGKGLTFDSGGYNIKTGPGCSIELMKFDMGGSAAVLGA 371
PP FIHL YKPPSG V KLALVGKGLTFDSGGYNIKTGPGC IELMKFDMGGSAAV GA
Sbjct: 181 PPHFIHLCYKPPSGPVNAKLALVGKGLTFDSGGYNIKTGPGCLIELMKFDMGGSAAVFGA 360
Query: 372 AKALGQIKPLGVEVH 386
AKALGQIKPLGVEVH
Sbjct: 361 AKALGQIKPLGVEVH 405
>BP046003
Length = 528
Score = 33.5 bits (75), Expect = 0.095
Identities = 19/39 (48%), Positives = 26/39 (65%), Gaps = 3/39 (7%)
Frame = -3
Query: 87 EKDLTRDAKSRFENS---ILNKIDLKLDGLLSEASSEED 122
E+ + K+ E+S ILN +D KL+GLL EASSEE+
Sbjct: 214 EEKKPEERKAHLESSSYIILNSLDSKLNGLLVEASSEEN 98
>TC9760
Length = 556
Score = 29.3 bits (64), Expect = 1.8
Identities = 21/73 (28%), Positives = 31/73 (41%)
Frame = -1
Query: 160 GLGEAVVAVAKSAQASNVAIVLASSEGLSSESKLSTAYAIASGAVLGLFEDQRYKSESKK 219
GL V+KSA AS ++ +ASS + S +I LG F+ +Y +
Sbjct: 367 GLPRTFTKVSKSAIASVKSLFMASSSSYAKVSAFLFTLSITLCLRLGKFDIAKYPPAEQI 188
Query: 220 PAVRSIDIIGLGT 232
P R I+ T
Sbjct: 187 PRARRSSIVVCST 149
>AV422403
Length = 465
Score = 27.3 bits (59), Expect = 6.8
Identities = 27/86 (31%), Positives = 35/86 (40%)
Frame = +1
Query: 55 HPTNIEAPKISFSATDVDVTEWKGDILAVGVTEKDLTRDAKSRFENSILNKIDLKLDGLL 114
H +IE KI VTEWK + +T +D+T D K S L+ + GL
Sbjct: 73 HQMDIETGKI--------VTEWKFEKDGADITMRDITNDTK----GSQLDPSESTFLGLD 216
Query: 115 SEASSEEDFSGKVGQSTVVRIKGLGS 140
+ D K G V I G GS
Sbjct: 217 DNRLCQWDMREKKGM--VQNIAGSGS 288
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.314 0.132 0.366
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,859,846
Number of Sequences: 28460
Number of extensions: 91641
Number of successful extensions: 320
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 319
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 319
length of query: 567
length of database: 4,897,600
effective HSP length: 95
effective length of query: 472
effective length of database: 2,193,900
effective search space: 1035520800
effective search space used: 1035520800
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC136953.9


