

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136953.3 + phase: 0 /pseudo
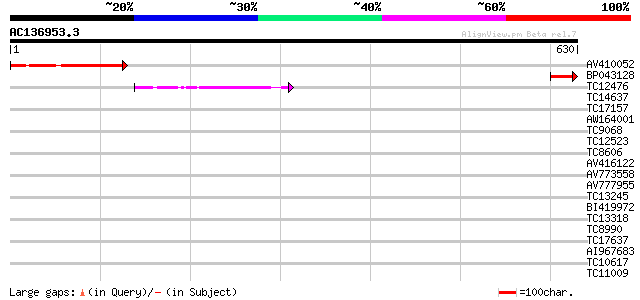
(630 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AV410052 207 5e-54
BP043128 61 5e-10
TC12476 similar to GB|AAC72109.1|3850569|F15K9 ESTs gb|T21276, g... 42 2e-04
TC14637 weakly similar to UP|FUS_HUMAN (P35637) RNA-binding prot... 36 0.021
TC17157 similar to UP|CAD27462 (CAD27462) Nucleosome assembly pr... 36 0.021
AW164001 35 0.036
TC9068 similar to UP|SCWA_YEAST (Q04951) Probable family 17 gluc... 35 0.036
TC12523 35 0.036
TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%) 35 0.048
AV416122 34 0.062
AV773558 34 0.062
AV777955 34 0.081
TC13245 similar to UP|Q7XIG8 (Q7XIG8) AcinusL protein-like, part... 33 0.11
BI419972 33 0.11
TC13318 similar to UP|CAA20314 (CAA20314) SPBC30B4.01c protein (... 33 0.14
TC8990 homologue to UP|SR52_LYCES (P49972) Signal recognition pa... 32 0.24
TC17637 32 0.31
AI967683 32 0.31
TC10617 similar to UP|Q9ARE3 (Q9ARE3) ZF-HD homeobox protein, pa... 32 0.40
TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Ar... 31 0.53
>AV410052
Length = 427
Score = 207 bits (526), Expect = 5e-54
Identities = 109/132 (82%), Positives = 119/132 (89%), Gaps = 1/132 (0%)
Frame = +3
Query: 1 MLEQLLIFTRGGLILWSCNEIGNALKGSPIDTLIRSCLLEERSGASSYNYDAPGAAYSLK 60
MLEQLLIFTRGGLILWS GNALKGSPIDTLIRSCLLEERSGA+SY +D+ YSLK
Sbjct: 48 MLEQLLIFTRGGLILWSW---GNALKGSPIDTLIRSCLLEERSGAASYTHDS----YSLK 206
Query: 61 WTFHNDLGLVFVAVYQRILHLLYVDDLLAAVKREFSQVYDPT-RTVYRDFDEIFKQLKIE 119
WTFHNDLGLVFVAVYQRILHLLYVDDLLAAVKREFS ++DP+ +T+Y DFDE F+QLKIE
Sbjct: 207 WTFHNDLGLVFVAVYQRILHLLYVDDLLAAVKREFSLIHDPSKKTLYPDFDETFRQLKIE 386
Query: 120 AEARAEDLKKSN 131
AEARAEDLKKS+
Sbjct: 387 AEARAEDLKKSS 422
>BP043128
Length = 384
Score = 61.2 bits (147), Expect = 5e-10
Identities = 29/29 (100%), Positives = 29/29 (100%)
Frame = -3
Query: 602 GAPVMFVGCGQSYTDLKKLNVKSIVKTLL 630
GAPVMFVGCGQSYTDLKKLNVKSIVKTLL
Sbjct: 382 GAPVMFVGCGQSYTDLKKLNVKSIVKTLL 296
>TC12476 similar to GB|AAC72109.1|3850569|F15K9 ESTs gb|T21276, gb|T45403,
and gb|AA586113 come from this gene. {Arabidopsis
thaliana;}, partial (7%)
Length = 791
Score = 42.4 bits (98), Expect = 2e-04
Identities = 50/180 (27%), Positives = 79/180 (43%), Gaps = 3/180 (1%)
Frame = +2
Query: 139 RKQQVTWKGDGSDGKKNGSAGGGLKNDGDGKNGKKNSEN-DRSAIVNNGNGYNLRSNGVV 197
R + T G G + K+ G ND G + ++ DR VNN N+ SN VV
Sbjct: 11 RAEVATGGGGGGENVKDLEVG----NDEKGVAAESETDGGDRLEEVNNDQ--NVVSN-VV 169
Query: 198 GNVSVNGKENDSVNNGAFDVNRLQKKVRNKGGNGKKTDAVVTKAEPKKVVKKNRVWDE-K 256
S +G++ D + N +V+ ++ N + K +D + +P +++ WDE +
Sbjct: 170 ATGSDSGEKLD-IKNEVKEVSEVKADHDNSESSCKDSDISIVSTQPSMPGEEDIGWDEIE 346
Query: 257 PVETKLDFTDHVD-IDGDADKDRKVDYLAKEQGESMMDKDEIFSSDSEDEEDDDDDNAGK 315
VE+ D VD ++ D + A E+ E + S D EDDDDD A K
Sbjct: 347 YVESHDDNKGGVDAVESSTSVDLRKRLSAAEEDEDL----------SWDIEDDDDDEAVK 496
>TC14637 weakly similar to UP|FUS_HUMAN (P35637) RNA-binding protein FUS
(Oncogene FUS) (Oncogene TLS) (Translocated in
liposarcoma protein) (POMp75) (75 kDa DNA-pairing
protein), partial (6%)
Length = 1332
Score = 35.8 bits (81), Expect = 0.021
Identities = 23/86 (26%), Positives = 41/86 (46%), Gaps = 2/86 (2%)
Frame = +2
Query: 256 KPVETKLDFTDHVDIDGDADKDRKVDYLAKEQGESMMDKDEIFSSDSED--EEDDDDDNA 313
+PV + D ++ KD +L K DKD ++ ED EE+D+DD++
Sbjct: 554 RPVTKFTEDFDFTAMNEKFKKDEVWGHLGKSNKSHSKDKDGEENAFDEDYNEEEDNDDSS 733
Query: 314 GKKSKPDAKKKGWFSSMFQSIAGKAN 339
+ KP K +F S+ +++ A+
Sbjct: 734 NAEVKPVYNKDDFFDSLSSNVSDNAS 811
>TC17157 similar to UP|CAD27462 (CAD27462) Nucleosome assembly protein
1-like protein 3, partial (81%)
Length = 1298
Score = 35.8 bits (81), Expect = 0.021
Identities = 29/130 (22%), Positives = 53/130 (40%), Gaps = 26/130 (20%)
Frame = +2
Query: 221 QKKVRNKGGNGKKTDAVVTKAE----------PKKVVKKNRVWDEKPVETKLDFTDHVDI 270
QK ++ K G K +TK E P +V + + D+ VE + +H
Sbjct: 506 QKILKKKPKKGSKNTKPITKTEKCESFFNFFNPPQVPEDDDDIDDDAVEELQNLMEHDYD 685
Query: 271 DGDADKDRKVDY----------------LAKEQGESMMDKDEIFSSDSEDEEDDDDDNAG 314
G +D+ + + + ++ + D+DE D E+E+DDDDD
Sbjct: 686 IGSTIRDKIIPHAVSWFTGEAEQSDFEDIEEDDEDGDEDEDEDDDDDEEEEDDDDDDEDD 865
Query: 315 KKSKPDAKKK 324
++ + +K K
Sbjct: 866 EEGEGKSKSK 895
Score = 33.9 bits (76), Expect = 0.081
Identities = 20/62 (32%), Positives = 28/62 (44%)
Frame = +2
Query: 263 DFTDHVDIDGDADKDRKVDYLAKEQGESMMDKDEIFSSDSEDEEDDDDDNAGKKSKPDAK 322
DF D + D D D+D D D DE D +D+EDD++ KSK +K
Sbjct: 758 DFEDIEEDDEDGDEDEDEDD----------DDDEEEEDDDDDDEDDEEGEGKSKSKSGSK 907
Query: 323 KK 324
+
Sbjct: 908 AR 913
>AW164001
Length = 447
Score = 35.0 bits (79), Expect = 0.036
Identities = 17/44 (38%), Positives = 26/44 (58%)
Frame = +1
Query: 269 DIDGDADKDRKVDYLAKEQGESMMDKDEIFSSDSEDEEDDDDDN 312
D+ GD+ + D + KE+ D DE F D +D++DDDDD+
Sbjct: 16 DVVGDSVNKQNDDEVYKEE-----DVDEDFDDDDDDDDDDDDDD 132
>TC9068 similar to UP|SCWA_YEAST (Q04951) Probable family 17 glucosidase
SCW10 precursor (Soluble cell wall protein 10) ,
partial (6%)
Length = 1209
Score = 35.0 bits (79), Expect = 0.036
Identities = 13/29 (44%), Positives = 20/29 (68%)
Frame = +2
Query: 293 DKDEIFSSDSEDEEDDDDDNAGKKSKPDA 321
D D+ D +D++DDDDD+ G K +PD+
Sbjct: 92 DDDDDDDDDDDDDDDDDDDDDGDKKQPDS 178
>TC12523
Length = 603
Score = 35.0 bits (79), Expect = 0.036
Identities = 24/73 (32%), Positives = 32/73 (42%), Gaps = 9/73 (12%)
Frame = +2
Query: 125 EDLKKSNPVIVGGNRKQQVTWKGDGSDGKKNGSAGGGLKNDGDGKNGKK---------NS 175
E+ K S PV GG+ + + G+ SD + G + G G+ GKK
Sbjct: 344 ENQKNSEPVGDGGSNEDCIDAAGEDSDHCDDDVCGDASGSGGGGEGGKKKKKIKKKKNKG 523
Query: 176 ENDRSAIVNNGNG 188
EN SA NNG G
Sbjct: 524 ENGGSAPPNNGGG 562
>TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%)
Length = 1008
Score = 34.7 bits (78), Expect = 0.048
Identities = 43/193 (22%), Positives = 64/193 (32%)
Frame = -2
Query: 131 NPVIVGGNRKQQVTWKGDGSDGKKNGSAGGGLKNDGDGKNGKKNSENDRSAIVNNGNGYN 190
N G K Q K ++G+ G +DGDG G+ E D S+ +G GY
Sbjct: 680 NMATTKGETKSQSEDKQHDAEGEDGNDDEGDEDDDGDGPFGE--GEEDLSS--EDGAGYG 513
Query: 191 LRSNGVVGNVSVNGKENDSVNNGAFDVNRLQKKVRNKGGNGKKTDAVVTKAEPKKVVKKN 250
SN N+ K +GG G
Sbjct: 512 NNSN-----------------------NKSNSKKAPEGGPGAGAG--------------- 447
Query: 251 RVWDEKPVETKLDFTDHVDIDGDADKDRKVDYLAKEQGESMMDKDEIFSSDSEDEEDDDD 310
P E D D D + D D D ++ G +++ + D+EDEE+D++
Sbjct: 446 ------PEENGEDEEDEDGEDQEDDDDDDEDDDEEDDGGEDEEEEGVEEEDNEDEEEDEE 285
Query: 311 DNAGKKSKPDAKK 323
D + KK
Sbjct: 284 DEEALQPPKKRKK 246
>AV416122
Length = 414
Score = 34.3 bits (77), Expect = 0.062
Identities = 18/52 (34%), Positives = 31/52 (59%)
Frame = +3
Query: 274 ADKDRKVDYLAKEQGESMMDKDEIFSSDSEDEEDDDDDNAGKKSKPDAKKKG 325
A+ D + D + +GE D +E + E+E+D+DDD+ G +++ D K KG
Sbjct: 258 ANDDNEPDNDVQFEGEEEEDDEEDKEEEDEEEDDEDDDSNG-EAEVDRKGKG 410
>AV773558
Length = 469
Score = 34.3 bits (77), Expect = 0.062
Identities = 16/50 (32%), Positives = 29/50 (58%), Gaps = 2/50 (4%)
Frame = -2
Query: 281 DYLAKEQGESMMDKDEI--FSSDSEDEEDDDDDNAGKKSKPDAKKKGWFS 328
D L +G+ +D D + +S D +D++DDDDD+ D ++ G++S
Sbjct: 429 DGLFFNEGDEELDDDFVLVYSEDDDDDDDDDDDDDDYTDDEDEEEGGFYS 280
>AV777955
Length = 429
Score = 33.9 bits (76), Expect = 0.081
Identities = 16/41 (39%), Positives = 25/41 (60%)
Frame = +1
Query: 296 EIFSSDSEDEEDDDDDNAGKKSKPDAKKKGWFSSMFQSIAG 336
E+ D EDE+D++DD +GK+ KP +K + W F + G
Sbjct: 43 EVAVEDEEDEDDEEDDESGKR-KP-SKPRSWTWDHFTKVIG 159
>TC13245 similar to UP|Q7XIG8 (Q7XIG8) AcinusL protein-like, partial (4%)
Length = 562
Score = 33.5 bits (75), Expect = 0.11
Identities = 22/71 (30%), Positives = 38/71 (52%)
Frame = -2
Query: 128 KKSNPVIVGGNRKQQVTWKGDGSDGKKNGSAGGGLKNDGDGKNGKKNSENDRSAIVNNGN 187
K+S+ V +G + T+ G G G+++ +AGG ++ G G G+ + N+ S++ G
Sbjct: 273 KRSSSVTIGRS-----TFSGGGGGGRRSLAAGGSFESGGGG--GRSIAGNEGSSLC*RGG 115
Query: 188 GYNLRSNGVVG 198
G L GV G
Sbjct: 114 GSGL-EEGVAG 85
>BI419972
Length = 498
Score = 33.5 bits (75), Expect = 0.11
Identities = 17/43 (39%), Positives = 24/43 (55%)
Frame = +1
Query: 278 RKVDYLAKEQGESMMDKDEIFSSDSEDEEDDDDDNAGKKSKPD 320
RK YLA + E+ D+D D +D+ DDDDD+ G+ D
Sbjct: 316 RKPAYLANKDSETEDDED---GDDEDDDADDDDDDEGEDYSGD 435
>TC13318 similar to UP|CAA20314 (CAA20314) SPBC30B4.01c protein (SPBC3D6.14C
protein) (Fragment), partial (15%)
Length = 543
Score = 33.1 bits (74), Expect = 0.14
Identities = 21/70 (30%), Positives = 34/70 (48%)
Frame = +3
Query: 256 KPVETKLDFTDHVDIDGDADKDRKVDYLAKEQGESMMDKDEIFSSDSEDEEDDDDDNAGK 315
KPV+T D + D D + D + + D D D+ D E+EEDDDDD+ +
Sbjct: 231 KPVQT--DNEEEEDDDEEEDDEDEDD-----------DDDDDDDDDGEEEEDDDDDDEEE 371
Query: 316 KSKPDAKKKG 325
+ + + +G
Sbjct: 372 EGSEEVEVEG 401
>TC8990 homologue to UP|SR52_LYCES (P49972) Signal recognition particle 54
kDa protein 2 (SRP54), partial (50%)
Length = 1141
Score = 32.3 bits (72), Expect = 0.24
Identities = 16/41 (39%), Positives = 24/41 (58%)
Frame = +3
Query: 590 KVGAALSMVYISGAPVMFVGCGQSYTDLKKLNVKSIVKTLL 630
K G ALS V + +PV+F+G G+ + + +VK V LL
Sbjct: 21 KGGGALSAVAATKSPVIFIGTGEHMDEFEVFDVKPFVSRLL 143
>TC17637
Length = 982
Score = 32.0 bits (71), Expect = 0.31
Identities = 39/146 (26%), Positives = 67/146 (45%), Gaps = 12/146 (8%)
Frame = +3
Query: 245 KVVKKNRVWDEKP--------VETKLDFTDHVDIDGDADKDRKVDYLAK---EQGESMMD 293
K+++ + D KP ++ L H++ D DA+ KVD L K E+ +S+ D
Sbjct: 27 KIIQPRKELDMKPKLELEIQQLKGSLSVLKHIEDDEDAEVLNKVDALHKDLREKEQSLQD 206
Query: 294 KDEIFSS-DSEDEEDDDDDNAGKKSKPDAKKKGWFSSMFQSIAGKANLEKSDLEPALKAL 352
DE+ + ++ + + +K D K+ +S I K + + D P L+A+
Sbjct: 207 IDELNQTLIVKERRSNTELQEARKELIDYIKE---TSSRGHIKVK-RMGELDTGPFLEAM 374
Query: 353 KDRLMTKNVAEEIAEKLCESVAASLE 378
K R + AEE A +LC A L+
Sbjct: 375 KKR-YNEEEAEERASELCSLWAEYLK 449
>AI967683
Length = 464
Score = 32.0 bits (71), Expect = 0.31
Identities = 23/81 (28%), Positives = 37/81 (45%)
Frame = +3
Query: 296 EIFSSDSEDEEDDDDDNAGKKSKPDAKKKGWFSSMFQSIAGKANLEKSDLEPALKALKDR 355
++++ D D+E +DDDN G + +SS + + G + E SD E + D
Sbjct: 54 KLYTQDISDDESEDDDNEGSAHDDSSA----YSSEPEELLGSS--ESSDSEETSDSDSD- 212
Query: 356 LMTKNVAEEIAEKLCESVAAS 376
+ EE+AE E V S
Sbjct: 213 -----IDEEVAEDYLEGVGGS 260
>TC10617 similar to UP|Q9ARE3 (Q9ARE3) ZF-HD homeobox protein, partial (16%)
Length = 622
Score = 31.6 bits (70), Expect = 0.40
Identities = 35/111 (31%), Positives = 46/111 (40%), Gaps = 7/111 (6%)
Frame = +2
Query: 111 EIFKQLKIEAEARAEDLKKSNPVIVGGNRKQQVTWKGDGSD--GKK-----NGSAGGGLK 163
E +++ + + R EDL VG +R W + + GK+ NG AGGG
Sbjct: 11 EFAERVGWKMQKRDEDLVMEFCHEVGVDRGVLKLWMHNHKNTFGKRDHAAANGGAGGG-- 184
Query: 164 NDGDGKNGKKNSENDRSAIVNNGNGYNLRSNGVVGNVSVNGKENDSVNNGA 214
D DG G + AI NGNG + N ENDS NGA
Sbjct: 185 -DDDGAGGGER------AITGNGNGS-------AASQDPNQYENDSGTNGA 295
>TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;}, partial (44%)
Length = 700
Score = 31.2 bits (69), Expect = 0.53
Identities = 17/63 (26%), Positives = 34/63 (52%), Gaps = 1/63 (1%)
Frame = +2
Query: 263 DFTDHVDIDGDADKDRKVDYLAKEQGESMMDKDEIFSSDSEDEEDDDDDN-AGKKSKPDA 321
D D D + + ++D +YL + + D++ + E++E DD+DN G+K+ +
Sbjct: 188 DDDDDDDEEEEEEEDLGTEYLVRRTVAAAEDEEASSDFEPEEDEGDDNDNDDGEKAGVPS 367
Query: 322 KKK 324
K+K
Sbjct: 368 KRK 376
Score = 29.3 bits (64), Expect = 2.0
Identities = 19/68 (27%), Positives = 26/68 (37%)
Frame = +2
Query: 259 ETKLDFTDHVDIDGDADKDRKVDYLAKEQGESMMDKDEIFSSDSEDEEDDDDDNAGKKSK 318
E DF D D D D GE + SD + DDD D+ G+ +
Sbjct: 284 EASSDFEPEEDEGDDNDND---------DGEKAGVPSKRKRSDKDGSGDDDSDDGGEDDE 436
Query: 319 PDAKKKGW 326
+K+ GW
Sbjct: 437 RPSKR*GW 460
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.314 0.133 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,286,995
Number of Sequences: 28460
Number of extensions: 93786
Number of successful extensions: 893
Number of sequences better than 10.0: 84
Number of HSP's better than 10.0 without gapping: 612
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 731
length of query: 630
length of database: 4,897,600
effective HSP length: 96
effective length of query: 534
effective length of database: 2,165,440
effective search space: 1156344960
effective search space used: 1156344960
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC136953.3


