

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136841.7 + phase: 0 /pseudo
(381 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
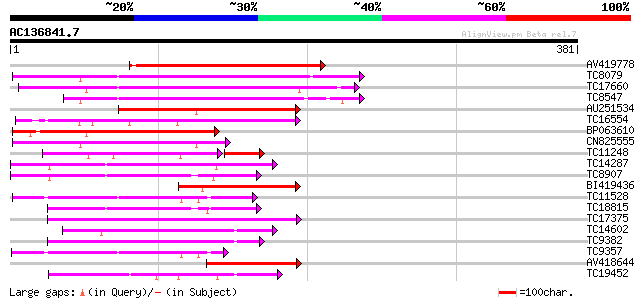
Score E
Sequences producing significant alignments: (bits) Value
AV419778 179 7e-46
TC8079 homologue to UP|Q9M6S1 (Q9M6S1) MAP kinase 3, complete 159 7e-40
TC17660 homologue to UP|Q9M6R8 (Q9M6R8) MAP kinase PsMAPK2, part... 141 2e-34
TC8547 homologue to UP|MMK1_MEDSA (Q07176) Mitogen-activated pro... 138 2e-33
AU251534 121 2e-28
TC16554 homologue to UP|AAN71719 (AAN71719) Glycogen synthase ki... 116 7e-27
BP063610 103 4e-23
CN825555 102 1e-22
TC11248 homologue to UP|P93323 (P93323) Cdc2MsF protein, partial... 77 1e-18
TC14287 similar to UP|Q9LEU7 (Q9LEU7) Serine/threonine protein k... 84 3e-17
TC8907 similar to UP|Q8H0X3 (Q8H0X3) Serine/threonine protein ki... 82 1e-16
BI419436 82 1e-16
TC11528 homologue to UP|Q39868 (Q39868) Protein kinase , partial... 80 4e-16
TC18815 similar to UP|Q8W1D5 (Q8W1D5) CBL-interacting protein ki... 80 6e-16
TC17375 mitogen-activated kinase kinase kinase alpha [Lotus corn... 80 6e-16
TC14602 UP|Q8GS56 (Q8GS56) Phosphoenolpyruvate carboxylase kinas... 78 3e-15
TC9382 weakly similar to UP|ABL_DROME (P00522) Tyrosine-protein ... 75 2e-14
TC9357 homologue to UP|Q39193 (Q39193) Protein kinase (Protein k... 72 1e-13
AV418644 70 4e-13
TC19452 homologue to UP|Q8S9J9 (Q8S9J9) At1g14000/F7A19_9, parti... 70 4e-13
>AV419778
Length = 411
Score = 179 bits (454), Expect = 7e-46
Identities = 82/132 (62%), Positives = 108/132 (81%)
Frame = +1
Query: 81 CFMKQMLQGLSHMHKKGFFHRDLKPENLLVTNDVLKIADFGLAREVSSMPPYTQYVSTRW 140
CF Q+ QGL++MH++G+FHRDLKPENLLVT D++K++DFGLARE+SS PPYT+YVSTRW
Sbjct: 10 CF--QVFQGLAYMHQRGYFHRDLKPENLLVTKDIIKVSDFGLAREISSHPPYTEYVSTRW 183
Query: 141 YRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESEIDQMYKIYCILGMPDSTCF 200
YRAPEVLLQS Y+ VDMWA+GAI+AELFTL P+FPG SE D++YKI ++G P + +
Sbjct: 184 YRAPEVLLQSYLYSSKVDMWAMGAIMAELFTLRPLFPGTSEADEIYKICSVIGSPTTESW 363
Query: 201 TIGANNSRLLDF 212
G +R +++
Sbjct: 364 ADGLQLARDINY 399
>TC8079 homologue to UP|Q9M6S1 (Q9M6S1) MAP kinase 3, complete
Length = 1550
Score = 159 bits (402), Expect = 7e-40
Identities = 93/244 (38%), Positives = 142/244 (58%), Gaps = 8/244 (3%)
Frame = +3
Query: 3 TFEIVAVKRLKRKFC-FWEEYTNLREIKALRKMNHQNIIKLREVV-----RENNELFFIF 56
T E+VAVK++ F + LREIK LR ++H+N+I LR+V+ RE +++
Sbjct: 357 TNELVAVKKIANAFDNHMDAKRTLREIKLLRHLDHENVIALRDVIPPPLRREFTDVYITT 536
Query: 57 EYMDCNLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTNDV-L 115
E MD +L+Q+I+ + SEE + F+ Q+L+GL ++H HRDLKP NLL+ ++ L
Sbjct: 537 ELMDTDLHQIIRSNQG-LSEEHCQYFLYQVLRGLKYIHSANIIHRDLKPSNLLLNSNCDL 713
Query: 116 KIADFGLAREVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPI 175
KI DFGLAR T+YV TRWYRAPE+LL S YT A+D+W++G I EL P+
Sbjct: 714 KIIDFGLARPTVENDFMTEYVVTRWYRAPELLLNSSDYTSAIDVWSVGCIFMELMNKKPL 893
Query: 176 FPGESEIDQMYKIYCILGMPDSTCFTIGANNSRLLDFVGHEVVAPVK-LSDIIPNASMEA 234
FPG+ + Q+ + +LG P + N + ++ P + L+ + P+ A
Sbjct: 894 FPGKDHVHQLRLLTELLGTPTEADLGL-VKNDDVRRYIRQLPQYPRQPLTKVFPHVHPMA 1070
Query: 235 IDLI 238
+DL+
Sbjct: 1071MDLV 1082
>TC17660 homologue to UP|Q9M6R8 (Q9M6R8) MAP kinase PsMAPK2, partial (64%)
Length = 717
Score = 141 bits (355), Expect = 2e-34
Identities = 88/239 (36%), Positives = 137/239 (56%), Gaps = 10/239 (4%)
Frame = +2
Query: 7 VAVKRLKRKF-CFWEEYTNLREIKALRKMNHQNIIKLREVVRENN-----ELFFIFEYMD 60
VA+K+++ F + LRE+K LR ++H N+I L++++ + +++ ++E MD
Sbjct: 8 VAIKKIQNAFENRVDALRTLRELKLLRHLHHDNVIALKDIMMPVHRSSFKDVYLVYELMD 187
Query: 61 CNLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVT-NDVLKIAD 119
+L+Q+IK + S + + F+ Q+L+GL ++H HRDLKP NLL+ N LKI D
Sbjct: 188 TDLHQIIKSSQS-LSNDHCQYFLFQLLRGLKYLHSANILHRDLKPGNLLINANCDLKICD 364
Query: 120 FGLAREVSSMPPY-TQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPG 178
FGLAR S + T+YV TRWYRAPE+LL Y ++D+W++G I AEL PIFPG
Sbjct: 365 FGLARINCSKNQFMTEYVVTRWYRAPELLLCCDNYGTSIDVWSVGCIFAELLGRKPIFPG 544
Query: 179 ESEIDQMYKIYCILG--MPDSTCFTIGANNSRLLDFVGHEVVAPVKLSDIIPNASMEAI 235
++Q+ I ILG + F + + + + + AP S + PNA AI
Sbjct: 545 SECLNQLKLIINILGSQREEDIEFIDNPKAKKYIKSLPYSIGAP--FSRLYPNAHPLAI 715
>TC8547 homologue to UP|MMK1_MEDSA (Q07176) Mitogen-activated protein
kinase homolog MMK1 (MAP kinase MSK7) (MAP kinase ERK1)
, partial (71%)
Length = 1098
Score = 138 bits (347), Expect = 2e-33
Identities = 80/211 (37%), Positives = 121/211 (56%), Gaps = 9/211 (4%)
Frame = +1
Query: 37 QNIIKLREVV----REN-NELFFIFEYMDCNLYQLIKEREKPFSEEEIRCFMKQMLQGLS 91
+N++ +R++V RE N+++ +E MD +L+Q+I+ + SEE + F+ Q+L+GL
Sbjct: 4 ENVVAIRDIVPPPQREVFNDVYIAYELMDTDLHQIIRSNQG-LSEEHCQYFLYQILRGLK 180
Query: 92 HMHKKGFFHRDLKPENLLVT-NDVLKIADFGLAREVSSMPPYTQYVSTRWYRAPEVLLQS 150
++H HRDLKP NLL+ N LKI DFGLAR S T+YV TRWYRAPE+LL S
Sbjct: 181 YIHSANVLHRDLKPSNLLLNANCDLKICDFGLARVTSETDFMTEYVVTRWYRAPELLLNS 360
Query: 151 PCYTPAVDMWAIGAILAELFTLTPIFPGESEIDQMYKIYCILGMPDSTCFTIGANNSRLL 210
YT A+D+W++G I EL P+FPG + Q+ + ++G P +G N
Sbjct: 361 SDYTAAIDVWSVGCIFMELMDRKPLFPGRDHVHQLRLLMELIGTPSED--DLGFLNENAK 534
Query: 211 DFVGHEVVAPVK---LSDIIPNASMEAIDLI 238
++ + P + + P EAIDL+
Sbjct: 535 RYI--RQLPPYRRQSFQEKFPQVHPEAIDLV 621
>AU251534
Length = 391
Score = 121 bits (304), Expect = 2e-28
Identities = 59/125 (47%), Positives = 82/125 (65%), Gaps = 3/125 (2%)
Frame = +2
Query: 74 FSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTND-VLKIADFGLAR--EVSSMP 130
F+E +++C+M+Q+L GL H H + HRD+K NLL+ N+ VLKIADFGLA + +
Sbjct: 5 FTEPQVKCYMRQLLSGLEHCHHRRVLHRDIKGSNLLIDNEGVLKIADFGLASVFDPNHKH 184
Query: 131 PYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESEIDQMYKIYC 190
P T V T WYR PE+LL + Y VD+W+ G IL EL PI PG +E++Q++KIY
Sbjct: 185 PMTSRVVTLWYRPPELLLGATDYDVGVDLWSAGCILGELLAGKPIMPGRTEVEQLHKIYK 364
Query: 191 ILGMP 195
+ G P
Sbjct: 365 LCGSP 379
>TC16554 homologue to UP|AAN71719 (AAN71719) Glycogen synthase kinase 3 beta
protein kinase DWARF12 , partial (71%)
Length = 927
Score = 116 bits (290), Expect = 7e-27
Identities = 68/202 (33%), Positives = 116/202 (56%), Gaps = 11/202 (5%)
Frame = +1
Query: 5 EIVAVKRLKRKFCFWEEYTNLREIKALRKMNHQNIIKLREV---VRENNELFF--IFEYM 59
E VA+K++ + Y N RE++ +R M+H N++ L+ +ELF + EY+
Sbjct: 310 EAVAIKKVLQD----RRYKN-RELQLMRVMDHPNVVSLKHCFFSTTSTDELFLNLVMEYV 474
Query: 60 DCNLYQLIKEREKPFSEEEI---RCFMKQMLQGLSHMHK-KGFFHRDLKPENLLVT--ND 113
++Y++IK I + +M Q+ +GL+++H G HRDLKP+N+LV
Sbjct: 475 PESMYRVIKHYSNANQRMPIIYVKLYMYQIFRGLAYIHTVPGVCHRDLKPQNILVDPLTH 654
Query: 114 VLKIADFGLAREVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLT 173
+K+ DFG A+ + Y+ +R+YRAPE++ + Y+ ++D+W+ G +LAEL
Sbjct: 655 QVKLCDFGSAKMLVKGEANISYICSRFYRAPELIFGATEYSTSIDIWSAGCVLAELLLGQ 834
Query: 174 PIFPGESEIDQMYKIYCILGMP 195
P+FPGE+ +DQ+ I +LG P
Sbjct: 835 PLFPGENAVDQLVHIIKVLGTP 900
>BP063610
Length = 461
Score = 103 bits (258), Expect = 4e-23
Identities = 62/146 (42%), Positives = 90/146 (61%), Gaps = 7/146 (4%)
Frame = -3
Query: 3 TFEIVAVKRL---KRKFCFWEEYTNLREIKALRKMNHQNIIKLREVVRENN--ELFFIFE 57
T EIVA+K++ K K F T+LREI L +H I+ ++EVV ++ +F + E
Sbjct: 459 TGEIVALKKVXMEKEKEGF--PLTSLREINILLSFHHPYIVDVKEVVVGSSLDSIFMVME 286
Query: 58 YMDCNLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTN-DVLK 116
YM+ +L L++ ++PFS+ E++C M Q+L+G+ ++H HRDLK NLL+ N LK
Sbjct: 285 YMEHDLKGLMESMKQPFSQSEVKCLMIQLLEGVKYLHDNWVLHRDLKTSNLLLNNRGELK 106
Query: 117 IADFGLAREVSS-MPPYTQYVSTRWY 141
I DFGLAR S + PYT V T WY
Sbjct: 105 ICDFGLARXYGSPLKPYTHLVVTLWY 28
>CN825555
Length = 714
Score = 102 bits (254), Expect = 1e-22
Identities = 62/152 (40%), Positives = 90/152 (58%), Gaps = 6/152 (3%)
Frame = +1
Query: 3 TFEIVAVKRLKRKFCFWEEYTNL-REIKALRKMNHQNIIKLREVV--RENNELFFIFEYM 59
T +IVA+K+++ E + REI LR+++H N++KL +V R + L+ +FEYM
Sbjct: 250 TGKIVALKKVRFDNLEPESVKFMAREILILRRLDHPNVLKLEGLVTSRMSCSLYLVFEYM 429
Query: 60 DCNLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTND-VLKIA 118
+L L F+E +++C+M Q+ GL H H + HRD+K NLL+ N+ VLKIA
Sbjct: 430 VHDLAGLATNPAIKFTESQVKCYMHQLFTGLEHCHNRHVLHRDIKGSNLLIDNEGVLKIA 609
Query: 119 DFGLAR--EVSSMPPYTQYVSTRWYRAPEVLL 148
DFGLA + P T V T WYR PE+LL
Sbjct: 610 DFGLASFFDPDHKHPMTSRVVTLWYRPPELLL 705
>TC11248 homologue to UP|P93323 (P93323) Cdc2MsF protein, partial (72%)
Length = 747
Score = 76.6 bits (187), Expect(2) = 1e-18
Identities = 46/133 (34%), Positives = 77/133 (57%), Gaps = 12/133 (9%)
Frame = +1
Query: 23 TNLREIKALRKMNHQ-NIIKLREVVRENNE-----LFFIFEYMDCNLYQLIK---EREKP 73
T LRE+ LR ++ ++++L +V + ++ L+ +FEYMD +L + I+ + +
Sbjct: 178 TTLREVSILRMLSRDPHVVRLMDVKQGQSKEGKTVLYLVFEYMDTDLKKFIRTFRQTGQN 357
Query: 74 FSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTND--VLKIADFGLAREVS-SMP 130
+ ++ M Q+ +G++ H G HRDLKP NLL+ + +LKIAD GLAR + +
Sbjct: 358 VPPKTVKSLMYQLRKGVAFCHGHGILHRDLKPHNLLMDRETNMLKIADLGLARAFTVPIK 537
Query: 131 PYTQYVSTRWYRA 143
YT + T WYRA
Sbjct: 538 KYTHEILTLWYRA 576
Score = 32.7 bits (73), Expect(2) = 1e-18
Identities = 15/27 (55%), Positives = 19/27 (69%)
Frame = +2
Query: 145 EVLLQSPCYTPAVDMWAIGAILAELFT 171
EVLL + Y+ AVDMW++ I AEL T
Sbjct: 581 EVLLGATHYSMAVDMWSVACIFAELVT 661
>TC14287 similar to UP|Q9LEU7 (Q9LEU7) Serine/threonine protein kinase-like
protein (CBL-interacting protein kinase 5), partial
(75%)
Length = 1999
Score = 84.3 bits (207), Expect = 3e-17
Identities = 57/186 (30%), Positives = 96/186 (50%), Gaps = 6/186 (3%)
Frame = +3
Query: 1 MRTFEIVAVKRLKRKFCFWEEYTNL--REIKALRKMNHQNIIKLREVVRENNELFFIFEY 58
+ T E VA+K +K++ + RE+ +R + H +I++L+EV+ ++F + EY
Sbjct: 492 LATNENVAIKVIKKEKLKKDRLVKQIKREVSVMRLVRHPHIVELKEVMATKGKIFLVMEY 671
Query: 59 MDCNLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLV-TNDVLKI 117
+ K + +E++ R + +Q++ + H +G HRDLKPENLL+ N+ LK+
Sbjct: 672 VKGGEL-FTKVNKGKLNEDDARKYFQQLISAVDFCHSRGVTHRDLKPENLLLDENEDLKV 848
Query: 118 ADFGLAREVSSMPPYTQYVS---TRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTP 174
+DFGL+ V+ T Y APEVL + D+W+ G IL L +
Sbjct: 849 SDFGLSALPEQRRDDGMLVTPCGTPAYVAPEVLKKKGYDGSKADIWSCGVILYALLSGYL 1028
Query: 175 IFPGES 180
F GE+
Sbjct: 1029PFQGEN 1046
>TC8907 similar to UP|Q8H0X3 (Q8H0X3) Serine/threonine protein kinase-like
protein, partial (44%)
Length = 941
Score = 82.4 bits (202), Expect = 1e-16
Identities = 58/179 (32%), Positives = 91/179 (50%), Gaps = 10/179 (5%)
Frame = +3
Query: 1 MRTFEIVAVKRLKRKFCFWEEYTNL--REIKALRKMNHQNIIKLREVVRENNELFFIFEY 58
M T E VA+K +K++ E RE+ +R + H +I++L+EV+ ++F + EY
Sbjct: 402 METNESVAIKVIKKERLKKERLVKQIKREVSVMRLVRHPHIVELKEVMATKTKIFMVVEY 581
Query: 59 MDCNLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLV-TNDVLKI 117
+ K + +E R + +Q++ + H +G HRDLKPENLL+ N+ LK+
Sbjct: 582 VKGGEL-FAKLTKGKMTEVAARKYFQQLISAVDFCHSRGVTHRDLKPENLLLDDNEDLKV 758
Query: 118 ADFGLAREVSSMPPYTQY-------VSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAEL 169
+DFGL SS+P + T Y APEVL + D+W+ G IL L
Sbjct: 759 SDFGL----SSLPEQRRSDGMLLTPCGTPAYVAPEVLKKKGYDGSKADIWSCGVILYAL 923
>BI419436
Length = 601
Score = 82.4 bits (202), Expect = 1e-16
Identities = 39/84 (46%), Positives = 55/84 (65%), Gaps = 2/84 (2%)
Frame = +1
Query: 114 VLKIADFGLAREVSS--MPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFT 171
VLK+ADFGLA SS P T V T WYR PE+LL + Y P+VD+W++G + AEL
Sbjct: 19 VLKVADFGLANFTSSGHKQPLTSRVVTLWYRPPELLLGATDYGPSVDLWSVGCVFAELLV 198
Query: 172 LTPIFPGESEIDQMYKIYCILGMP 195
P+ G +E++Q++KI+ + G P
Sbjct: 199 GKPVLQGRTEVEQLHKIFKLCGSP 270
>TC11528 homologue to UP|Q39868 (Q39868) Protein kinase , partial (56%)
Length = 640
Score = 80.5 bits (197), Expect = 4e-16
Identities = 62/170 (36%), Positives = 87/170 (50%), Gaps = 6/170 (3%)
Frame = +3
Query: 3 TFEIVAVKRLKRKFCFWEEYTNLREIKALRKMNHQNIIKLREVVRENNELFFIFEYMDCN 62
T E+VAVK ++R E REI R + H NII+ +EV+ L + EY
Sbjct: 144 TGELVAVKYIERGKKIDENVQ--REIINHRSLRHPNIIRFKEVLLTPTHLAIVLEYASGG 317
Query: 63 -LYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTNDV---LKIA 118
L++ I + FSE+E R F +Q++ G+S+ H HRDLK EN L+ + LKI
Sbjct: 318 ELFERICSAGR-FSEDEARYFFQQLISGVSYCHSMEICHRDLKLENTLLDGNPSPRLKIC 494
Query: 119 DFGLARE--VSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAIL 166
DFG ++ + S P T V T Y APEVL + D+W+ G L
Sbjct: 495 DFGYSKSAILHSQPKST--VGTPAYIAPEVLSRKEYDGKVADVWSCGVTL 638
>TC18815 similar to UP|Q8W1D5 (Q8W1D5) CBL-interacting protein kinase
CIPK25, partial (58%)
Length = 979
Score = 80.1 bits (196), Expect = 6e-16
Identities = 54/153 (35%), Positives = 80/153 (51%), Gaps = 9/153 (5%)
Frame = +2
Query: 26 REIKALRKMNHQNIIKLREVVRENNELFFIFEYMDCNLYQLIKEREKPFSEEEIRCFMKQ 85
REI +R + H NI+ L+EV+ ++FFI EY+ K + ++ R + +Q
Sbjct: 179 REISIMRLVRHPNIVNLKEVMATKTKIFFIMEYIRGGEL-FAKVAKGKLKDDLARRYFQQ 355
Query: 86 MLQGLSHMHKKGFFHRDLKPENLLV-TNDVLKIADFGLAREVSSMPP--------YTQYV 136
++ + + H +G HRDLKPENLL+ N+ LK++DFGL S +P +TQ
Sbjct: 356 LISAVDYCHSRGVSHRDLKPENLLLDENENLKVSDFGL----SGLPEQLRQDGLLHTQ-C 520
Query: 137 STRWYRAPEVLLQSPCYTPAVDMWAIGAILAEL 169
T Y APEVL + D W+ G IL L
Sbjct: 521 GTPAYVAPEVLRKKGYDGFKTDTWSCGVILYAL 619
>TC17375 mitogen-activated kinase kinase kinase alpha [Lotus corniculatus
var. japonicus]
Length = 1422
Score = 80.1 bits (196), Expect = 6e-16
Identities = 45/172 (26%), Positives = 88/172 (51%), Gaps = 1/172 (0%)
Frame = +3
Query: 26 REIKALRKMNHQNIIKLREVVRENNELFFIFEYMDCNLYQLIKEREKPFSEEEIRCFMKQ 85
+EI L + +H NI++ L EY+ + + F E I+ + +Q
Sbjct: 78 QEINLLNQFSHPNIVQYYGSELGEESLSVYLEYVSGGSIHKLLQEYGAFKEPVIQNYTRQ 257
Query: 86 MLQGLSHMHKKGFFHRDLKPENLLV-TNDVLKIADFGLAREVSSMPPYTQYVSTRWYRAP 144
++ GL+++H + HRD+K N+LV N +K+ADFG+++ ++S + + ++ AP
Sbjct: 258 IVSGLAYLHSRNTVHRDIKGANILVDPNGEIKLADFGMSKHINSAASMLSFKGSPYWMAP 437
Query: 145 EVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESEIDQMYKIYCILGMPD 196
EV++ + Y VD+ ++G + E+ T P + + ++KI MP+
Sbjct: 438 EVVMNTNGYGLPVDISSLGCTILEMATSKPPWSQFEGVAAIFKIGNSKDMPE 593
>TC14602 UP|Q8GS56 (Q8GS56) Phosphoenolpyruvate carboxylase kinase, complete
Length = 1256
Score = 77.8 bits (190), Expect = 3e-15
Identities = 50/148 (33%), Positives = 78/148 (51%), Gaps = 3/148 (2%)
Frame = +2
Query: 36 HQNIIKLREVVRENNELFFIFEYMD--CNLYQLIKEREKPFSEEEIRCFMKQMLQGLSHM 93
H NI+++ +V +++ L + E L +++ E E MKQ+L+ ++H
Sbjct: 242 HPNILQIFDVFEDDDVLSMVIELCQPLTLLDRIVAANGTSIPEVEAAGLMKQLLEAVAHC 421
Query: 94 HKKGFFHRDLKPENLLV-TNDVLKIADFGLAREVSSMPPYTQYVSTRWYRAPEVLLQSPC 152
H+ G HRD+KP+N+L LK+ADFG A + V T +Y APEVL+
Sbjct: 422 HRLGVAHRDVKPDNVLFGGGGDLKLADFGSAEWFGDGRRMSGVVGTPYYVAPEVLMGRE- 598
Query: 153 YTPAVDMWAIGAILAELFTLTPIFPGES 180
Y VD+W+ G IL + + TP F G+S
Sbjct: 599 YGEKVDVWSCGVILYIMLSGTPPFYGDS 682
>TC9382 weakly similar to UP|ABL_DROME (P00522) Tyrosine-protein kinase Abl
(D-ash) , partial (5%)
Length = 1197
Score = 74.7 bits (182), Expect = 2e-14
Identities = 43/148 (29%), Positives = 78/148 (52%), Gaps = 2/148 (1%)
Frame = +2
Query: 26 REIKALRKMNHQNIIKLREVVRENNELFFIFEYMDC-NLYQLIKEREKPFSEEEIRCFMK 84
+E+ +RK+ H+N+++ L + E+M +LY + ++ F +
Sbjct: 77 QEVYIMRKIRHKNVVQFIGACTRTPNLCIVTEFMSRGSLYDFLHKQRGVFKLPSLLKVAI 256
Query: 85 QMLQGLSHMHKKGFFHRDLKPENLLVT-NDVLKIADFGLAREVSSMPPYTQYVSTRWYRA 143
+ +G++++H+ HRDLK NLL+ N+++K+ADFG+AR ++ T T + A
Sbjct: 257 DVSKGMNYLHQNNIIHRDLKTGNLLMDENELVKVADFGVARVITQSGVMTAETGTYRWMA 436
Query: 144 PEVLLQSPCYTPAVDMWAIGAILAELFT 171
PEV+ P Y D+++ G L EL T
Sbjct: 437 PEVIEHKP-YDQKADVFSFGIALWELLT 517
>TC9357 homologue to UP|Q39193 (Q39193) Protein kinase (Protein kinase,
41K) , partial (71%)
Length = 1181
Score = 72.4 bits (176), Expect = 1e-13
Identities = 56/152 (36%), Positives = 80/152 (51%), Gaps = 6/152 (3%)
Frame = +1
Query: 2 RTFEIVAVKRLKRKFCFWEEYTNLREIKALRKMNHQNIIKLREVVRENNELFFIFEYMD- 60
+T E+VAVK ++R E REI R + H NI++ +EV+ L + EY
Sbjct: 424 QTKELVAVKYIERGDKIDENVK--REIINHRSLRHPNIVRFKEVILTPTHLAIVMEYASG 597
Query: 61 CNLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTNDV---LKI 117
L++ I + F+E+E R F +Q++ G+S+ H HRDLK EN L+ LKI
Sbjct: 598 GELFERICNAGR-FTEDEARFFFQQLISGVSYCHAMQVCHRDLKLENTLLDGSPTPRLKI 774
Query: 118 ADFGLARE--VSSMPPYTQYVSTRWYRAPEVL 147
DFG ++ + S P T V T Y APEVL
Sbjct: 775 CDFGYSKSSVLHSQPKST--VGTPAYIAPEVL 864
>AV418644
Length = 355
Score = 70.5 bits (171), Expect = 4e-13
Identities = 32/64 (50%), Positives = 44/64 (68%)
Frame = +3
Query: 133 TQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESEIDQMYKIYCIL 192
T V TRW+RAPE+L S Y +D+W++G I AEL TL P+FPG ++IDQ+ +I +L
Sbjct: 114 TSCVGTRWFRAPELLYGSTNYGFEIDLWSLGCIFAELLTLKPLFPGTADIDQLSRIINVL 293
Query: 193 GMPD 196
G D
Sbjct: 294 GNLD 305
>TC19452 homologue to UP|Q8S9J9 (Q8S9J9) At1g14000/F7A19_9, partial (41%)
Length = 562
Score = 70.5 bits (171), Expect = 4e-13
Identities = 52/168 (30%), Positives = 81/168 (47%), Gaps = 11/168 (6%)
Frame = +3
Query: 27 EIKALRKMNHQNIIKLREVVRENNELFFIFEYM-DCNLYQLIKEREKPFSEEEIRCFMKQ 85
E+ L K+ H NI++ V E L I EY+ +L+Q +KE+ I F
Sbjct: 48 EVNLLVKLRHPNIVQFLGAVTERKPLMLITEYLRGGDLHQYLKEKGSLSPSTAIN-FSMD 224
Query: 86 MLQGLSHMHKKG--FFHRDLKPENLLVTN---DVLKIADFGLAREVSSMPPYTQYVST-- 138
+++G++++H + HRDLKP N+L+ N D LK+ DFGL++ ++ + Y T
Sbjct: 225 IVRGMAYLHNEPNVIIHRDLKPRNVLLVNSSADHLKVGDFGLSKLITVQNSHDVYKMTGE 404
Query: 139 ---RWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESEID 183
Y APEV Y VD+++ IL E+ P F D
Sbjct: 405 TGSYRYMAPEVFKHRK-YDKKVDVYSFAMILYEMLEGEPPFASYEPYD 545
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.326 0.140 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,805,736
Number of Sequences: 28460
Number of extensions: 90063
Number of successful extensions: 731
Number of sequences better than 10.0: 148
Number of HSP's better than 10.0 without gapping: 667
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 674
length of query: 381
length of database: 4,897,600
effective HSP length: 92
effective length of query: 289
effective length of database: 2,279,280
effective search space: 658711920
effective search space used: 658711920
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 56 (26.2 bits)
Medicago: description of AC136841.7


