

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136841.5 + phase: 0
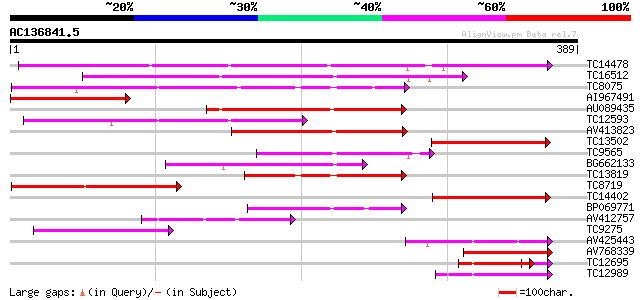
(389 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC14478 weakly similar to UP|Q94G86 (Q94G86) Beta-1,3-glucanase-... 239 7e-64
TC16512 similar to UP|Q9FXL4 (Q9FXL4) Elicitor inducible chitina... 208 1e-54
TC8075 similar to UP|E13B_PEA (Q03467) Glucan endo-1,3-beta-gluc... 156 5e-39
AI967491 141 2e-34
AU089435 132 1e-31
TC12593 131 2e-31
AV413823 110 3e-25
TC13502 98 3e-21
TC9565 similar to UP|Q9MAQ2 (Q9MAQ2) CDS, partial (19%) 96 8e-21
BG662133 93 9e-20
TC13819 similar to UP|Q9ZP12 (Q9ZP12) Glucan endo-1,3-beta-d-glu... 88 2e-18
TC8719 similar to UP|Q8H0I0 (Q8H0I0) Beta-1,3-glucanase-like pro... 84 4e-17
TC14402 similar to UP|Q8L9D9 (Q8L9D9) Beta-1,3-glucanase-like pr... 79 1e-15
BP069771 76 1e-14
AV412757 74 3e-14
TC9275 similar to UP|Q9M4A9 (Q9M4A9) Beta-1,3 glucanase precurso... 72 1e-13
AV425443 72 2e-13
AV768339 65 1e-11
TC12695 similar to UP|Q9SAK1 (Q9SAK1) T8K14.10, partial (17%) 60 1e-10
TC12989 similar to GB|AAO39944.1|28372924|BT003716 At5g08000 {Ar... 61 3e-10
>TC14478 weakly similar to UP|Q94G86 (Q94G86) Beta-1,3-glucanase-like
protein, partial (83%)
Length = 1784
Score = 239 bits (609), Expect = 7e-64
Identities = 144/407 (35%), Positives = 220/407 (53%), Gaps = 41/407 (10%)
Frame = +1
Query: 7 VTNVSSNQTLANQWVQTNLVPFYSKTLIRYLLVGNELISSTTNQTWPHIVPAMYRMKHSL 66
+ N++++ A QWV +N++P+Y + I + VGNE+++S +VPA+ ++ +L
Sbjct: 319 IPNLAADPNAAVQWVNSNVLPYYPASNITLITVGNEVLTSGDPNLASQLVPAIRNVQSAL 498
Query: 67 TIFGLH-KVKVGTPLAMDVLQTSFPPSNGTFRNDIALSVMKPMLEFLHVTNSFFFLDVYP 125
T L K+KV T +M VL S PPS+G+F N +K +L+FL +S F ++ YP
Sbjct: 499 TSASLGGKIKVSTVQSMAVLSQSDPPSSGSF-NAAFQDNLKQLLQFLKDNHSPFTINPYP 675
Query: 126 FFAWTSDPININLDYALFESDNITVTDSGTGLVYTNLFDQMVDAVYFAMERLGYPDIQIF 185
FFA+ SDP L + LF+ + V DSG G +YTN+FD VDAVY A+ LG+ I+I
Sbjct: 676 FFAYQSDPRPETLAFCLFQPNKGRV-DSGNGKLYTNMFDAQVDAVYSALSSLGFQGIEIV 852
Query: 186 IAETGWPNDGDLDQIGANIHNAGTYNRNFVKKVTKKPPVGTPARPGSILPSFIFALYNEN 245
+AETGWP+ GD +++G ++ NA YN N + + + VGTP PG + ++IFA+Y+E+
Sbjct: 853 VAETGWPSRGDSNEVGPSVDNAKAYNGNLITHL--RSLVGTPLMPGKSVDTYIFAIYDED 1026
Query: 246 LKTGLGTERHFGLLYPNGSRIYEIDL--SG-KTPEYEYKPLPPPDDYKGKAWCV------ 296
LK G G+ER FGL + + Y++ L SG +TP P P G +WCV
Sbjct: 1027LKPGKGSERAFGLFKTDLTMSYDVGLFKSGQQTPTTPVTPAPS----AGASWCVAKQGVS 1194
Query: 297 -------------------------------VAEGANKTAVVEALSYACSQGNRTCELVQ 325
+ +GA+ + L Y CSQ N C +Q
Sbjct: 1195DAQLQAHLDYVCSQSHIDCGPIPVQELHGVNLKQGASDAQLQANLDYVCSQSNIDCGPIQ 1374
Query: 326 PGKPCFEPDSVVGHASYAFSSYWAQFRRVGGTCNFNGLATQIAEDPS 372
G CFEPD+V HAS+A + Y+ R C+F+ ++++PS
Sbjct: 1375SGGACFEPDTVASHASFAMNLYYQTSGRNSWNCDFSQTGMLVSQNPS 1515
>TC16512 similar to UP|Q9FXL4 (Q9FXL4) Elicitor inducible chitinase
Nt-SubE76, partial (48%)
Length = 857
Score = 208 bits (530), Expect = 1e-54
Identities = 116/284 (40%), Positives = 162/284 (56%), Gaps = 20/284 (7%)
Frame = +2
Query: 51 TWPHIVPAMYRMKHSLTIFGLHK-VKVGTPLAMDVLQTSFPPSNGTFRNDIALSVMKPML 109
T P +VPAM + +L LH +KV +P+A+ L S+P S+G+FR ++ SV KPML
Sbjct: 11 TTPFLVPAMRNIHQALVKHNLHHDIKVSSPIALSALGNSYPSSSGSFRPELVQSVFKPML 190
Query: 110 EFLHVTNSFFFLDVYPFFAWTSDPININLDYALFESDNITVTDSGTGLVYTNLFDQMVDA 169
EFL T S+ ++VYPFFA+ S+ I LDYALF N V D G GL Y NLFD +DA
Sbjct: 191 EFLRETGSYLMVNVYPFFAYESNADVIPLDYALFR-QNPGVADPGNGLRYFNLFDAQIDA 367
Query: 170 VYFAMERLGYPDIQIFIAETGWPNDGDLDQIGANIHNAGTYNRNFVKKVTKKPPVGTPAR 229
V+ A+ L + D+ I ++ETGWP+ GD +++GA+ NA YN V+K+ GTP R
Sbjct: 368 VFAALSALKFNDVNIVVSETGWPSKGDENELGASAENAAAYNGGLVRKILTNS--GTPMR 541
Query: 230 PGSILPSFIFALYNENLKTGLGTERHFGLLYPNGSRIYEIDLS-----------GKTPEY 278
P + L ++FAL+NEN K G +ER+FGL YP+ ++Y+I L+
Sbjct: 542 PKADLTVYLFALFNENQKPGPTSERNFGLFYPDQKKVYDIPLTV*GLKSYHDKPSPVGGG 721
Query: 279 EYKPLPPP--------DDYKGKAWCVVAEGANKTAVVEALSYAC 314
E +P+P P G WCV A K + AL +AC
Sbjct: 722 EKQPVPSPAPVSGGVSKSTTGNNWCVANPDAEKVKLQAALDFAC 853
>TC8075 similar to UP|E13B_PEA (Q03467) Glucan endo-1,3-beta-glucosidase
precursor ((1->3)-beta-glucan endohydrolase)
((1->3)-beta-glucanase) (Beta-1,3-endoglucanase) ,
partial (88%)
Length = 1437
Score = 156 bits (395), Expect = 5e-39
Identities = 90/277 (32%), Positives = 154/277 (55%), Gaps = 4/277 (1%)
Frame = +2
Query: 2 LPNELVTNVSSNQTLANQWVQTNLVPFYSKTLIRYLLVGNELI---SSTTNQTWPHIVPA 58
+PN + ++++N A WVQ N++ F+ I+Y+ VGNE+ + T +++PA
Sbjct: 356 VPNSDLQSLATNADNARNWVQRNVLNFWPSVRIKYIAVGNEVSPVGGAPTQWMAQYVLPA 535
Query: 59 MYRMKHSLTIFGLH-KVKVGTPLAMDVLQTSFPPSNGTFRNDIALSVMKPMLEFLHVTNS 117
+ ++ GLH ++KV T + ++ S+PPS G+FR+D+ S + P + +L +
Sbjct: 536 TQNIYQAIRAQGLHDQIKVTTAIDTTLIGNSYPPSQGSFRSDVR-SYLDPFIGYLVYAGA 712
Query: 118 FFFLDVYPFFAWTSDPININLDYALFESDNITVTDSGTGLVYTNLFDQMVDAVYFAMERL 177
++VYP+F+ +P +++L YALF S NI D G Y NLFD M+DAV+ A++
Sbjct: 713 PLLVNVYPYFSHIGNPRDVSLSYALFTSPNIVAQDGQYG--YQNLFDAMLDAVHAAIDNT 886
Query: 178 GYPDIQIFIAETGWPNDGDLDQIGANIHNAGTYNRNFVKKVTKKPPVGTPARPGSILPSF 237
+++ ++E+GWP+DG + NA Y N ++ V + GTP RP ++
Sbjct: 887 KIGYVEVVVSESGWPSDGG---SATSYDNARIYLDNLIRHVGR----GTPRRPNKPTETY 1045
Query: 238 IFALYNENLKTGLGTERHFGLLYPNGSRIYEIDLSGK 274
IFA+++EN K+ E+HFGL N + Y G+
Sbjct: 1046IFAMFDENQKSP-EYEKHFGLFSANKQKKYPFGFGGE 1153
>AI967491
Length = 385
Score = 141 bits (355), Expect = 2e-34
Identities = 64/83 (77%), Positives = 76/83 (91%)
Frame = +3
Query: 1 MLPNELVTNVSSNQTLANQWVQTNLVPFYSKTLIRYLLVGNELISSTTNQTWPHIVPAMY 60
M+PNELVTN+S+NQTL++ W+Q+N+VPFY KTLIRYLLVGNEL SST N+TWPHIVPAM
Sbjct: 132 MVPNELVTNISTNQTLSDHWIQSNVVPFYRKTLIRYLLVGNELTSSTGNETWPHIVPAMR 311
Query: 61 RMKHSLTIFGLHKVKVGTPLAMD 83
R+KHSLTIF LHK+KVGTP AM+
Sbjct: 312 RIKHSLTIFHLHKIKVGTPFAME 380
>AU089435
Length = 423
Score = 132 bits (332), Expect = 1e-31
Identities = 65/137 (47%), Positives = 91/137 (65%)
Frame = +2
Query: 136 INLDYALFESDNITVTDSGTGLVYTNLFDQMVDAVYFAMERLGYPDIQIFIAETGWPNDG 195
I++DYALF+ +N V DSG GL YTNLFD +DAV+ AM L Y D+++ + ETGW + G
Sbjct: 17 ISIDYALFK-ENPGVVDSGNGLKYTNLFDAQLDAVFAAMSALKYDDVKVAVTETGWLSSG 193
Query: 196 DLDQIGANIHNAGTYNRNFVKKVTKKPPVGTPARPGSILPSFIFALYNENLKTGLGTERH 255
D +++GA NA YN N ++V GTP RP L F+FAL+NEN K G +ER+
Sbjct: 194 DSNEVGAGQENAAAYNGNLARRVLS--GAGTPLRPKEALDVFLFALFNENQKPGPTSERN 367
Query: 256 FGLLYPNGSRIYEIDLS 272
+GL YP+ ++Y+I L+
Sbjct: 368 YGLFYPSEKKVYDIPLT 418
>TC12593
Length = 846
Score = 131 bits (329), Expect = 2e-31
Identities = 75/198 (37%), Positives = 120/198 (59%), Gaps = 3/198 (1%)
Frame = +1
Query: 10 VSSNQTLANQWVQTNLVPFYSKTLIRYLLVGNELISSTTNQTWPHIVPAMYRMKHSLTI- 68
++S+ A WV +N+ P+Y + I + VGNE+I+S ++PA+ ++++L
Sbjct: 259 LASDPNFAKTWVSSNVAPYYPASNIILITVGNEVITSNDTGLVNQMLPAIQNVQNALDAA 438
Query: 69 -FGLHKVKVGTPLAMDVLQTSFPPSNGTFRNDIALSVMKPMLEFLHVTNSFFFLDVYPFF 127
FG K+KV T +M VL S PPS G F + +V++ +L F + T S F ++ YP+F
Sbjct: 439 SFGGGKIKVSTVHSMAVLGNSEPPSGGKFHAEYD-TVLQGLLSFNNATGSPFAINPYPYF 615
Query: 128 AWTSDPINI-NLDYALFESDNITVTDSGTGLVYTNLFDQMVDAVYFAMERLGYPDIQIFI 186
A+ SDP NL + LF+ ++ V D T L Y N+FD VDAV A++ LG+ +++I +
Sbjct: 616 AYKSDPGRPDNLAFCLFQPNSGRV-DPNTNLNYMNMFDAQVDAVRSALDSLGFKNVEIVV 792
Query: 187 AETGWPNDGDLDQIGANI 204
AETGWP GD D++G ++
Sbjct: 793 AETGWPYKGDNDEVGPSL 846
>AV413823
Length = 396
Score = 110 bits (276), Expect = 3e-25
Identities = 51/121 (42%), Positives = 78/121 (64%)
Frame = +2
Query: 153 SGTGLVYTNLFDQMVDAVYFAMERLGYPDIQIFIAETGWPNDGDLDQIGANIHNAGTYNR 212
+ T L YTN+FD +VDA YFAM L + +I + + E+GWP+ GD + A I NA TYN
Sbjct: 2 ANTLLHYTNVFDAIVDAAYFAMSDLNFTNIPVLVTESGWPSKGDSSEPDATIDNANTYNS 181
Query: 213 NFVKKVTKKPPVGTPARPGSILPSFIFALYNENLKTGLGTERHFGLLYPNGSRIYEIDLS 272
N ++ V GTP PG + ++I+ LYNE+L++G +E+++GL + NG +Y + L+
Sbjct: 182 NLIRHVLNN--TGTPKHPGIAVSTYIYELYNEDLRSGPVSEKNWGLFFANGGPVYTLHLT 355
Query: 273 G 273
G
Sbjct: 356 G 358
>TC13502
Length = 517
Score = 97.8 bits (242), Expect = 3e-21
Identities = 43/82 (52%), Positives = 53/82 (64%)
Frame = +2
Query: 290 KGKAWCVVAEGANKTAVVEALSYACSQGNRTCELVQPGKPCFEPDSVVGHASYAFSSYWA 349
KGK WC + AN + ALS AC++GN C+ + PGK C EP S+ HASY FSSYW+
Sbjct: 56 KGKVWCXGPKEANLMELAAALSNACNEGNIACDALAPGKECHEPASITDHASYVFSSYWS 235
Query: 350 QFRRVGGTCNFNGLATQIAEDP 371
+FR G +C FNGLA Q DP
Sbjct: 236 RFRSEGASCYFNGLAKQTTPDP 301
>TC9565 similar to UP|Q9MAQ2 (Q9MAQ2) CDS, partial (19%)
Length = 872
Score = 96.3 bits (238), Expect = 8e-21
Identities = 55/142 (38%), Positives = 75/142 (52%), Gaps = 20/142 (14%)
Frame = +3
Query: 170 VYFAMERLGYPDIQIFIAETGWPNDGDLDQIGANIHNAGTYNRNFVKKVTKKPPVGTPAR 229
VY A+ RLG + + I+ETGWP+ GD Q GA + NA YN N +K +KK GTP +
Sbjct: 15 VYAALSRLGCAKVPVHISETGWPSKGDDHQAGATVENAKRYNGNVIKISSKK---GTPLK 185
Query: 230 PGSILPSFIFALYNENLKTGLGTERHFGLLYPNGSRIYEIDLS----------------- 272
P + L ++FAL+NEN K G +ER++GL P+G+ Y + LS
Sbjct: 186 PDADLNIYVFALFNENQKPGPASERNYGLFKPDGTLAYSLGLSPSSFSSPSPSTNATTGG 365
Query: 273 ---GKTPEYEYKPLPPPDDYKG 291
G+ P PLPPP G
Sbjct: 366 GGGGRAP-----PLPPPTSSSG 416
>BG662133
Length = 428
Score = 92.8 bits (229), Expect = 9e-20
Identities = 47/140 (33%), Positives = 80/140 (56%), Gaps = 2/140 (1%)
Frame = +3
Query: 108 MLEFLHVTNSFFFLDVYPFFAWTSDPININLDYALFES--DNITVTDSGTGLVYTNLFDQ 165
+L+FL TNS + L+ YP++ +T ++YALF + D T Y ++FD
Sbjct: 15 LLQFLKNTNSSYMLNAYPYYGYTKGDGIFPIEYALFRPLPSVKQIVDPNTLYHYNSMFDA 194
Query: 166 MVDAVYFAMERLGYPDIQIFIAETGWPNDGDLDQIGANIHNAGTYNRNFVKKVTKKPPVG 225
MVDA Y++++ L + I I + ETGWP+ G ++ A NA TY+ N +++V G
Sbjct: 195 MVDATYYSIDALNFKGIPIVVTETGWPSFGGANEPDATEKNAETYSNNMIRRVMNDS--G 368
Query: 226 TPARPGSILPSFIFALYNEN 245
P++P + ++I+ L+NE+
Sbjct: 369 PPSQPSIPINTYIYELFNED 428
>TC13819 similar to UP|Q9ZP12 (Q9ZP12) Glucan endo-1,3-beta-d-glucosidase
precursor , partial (31%)
Length = 485
Score = 88.2 bits (217), Expect = 2e-18
Identities = 43/111 (38%), Positives = 71/111 (63%)
Frame = +3
Query: 162 LFDQMVDAVYFAMERLGYPDIQIFIAETGWPNDGDLDQIGANIHNAGTYNRNFVKKVTKK 221
LFD ++D VY A+E+ G PD+++ ++E+GWP+ G AN+ NA +Y +N ++ V
Sbjct: 3 LFDAILDGVYAALEKAGAPDMKVVVSESGWPSAGG---DAANVQNAESYYKNLIQHVKG- 170
Query: 222 PPVGTPARPGSILPSFIFALYNENLKTGLGTERHFGLLYPNGSRIYEIDLS 272
GTP RP + +++FA+++EN K TER+FGL P+ S Y+I+ +
Sbjct: 171 ---GTPKRPNGPIETYLFAMFDENRKPDPETERNFGLFRPDKSAKYQINFN 314
>TC8719 similar to UP|Q8H0I0 (Q8H0I0) Beta-1,3-glucanase-like protein,
partial (48%)
Length = 643
Score = 84.0 bits (206), Expect = 4e-17
Identities = 43/118 (36%), Positives = 73/118 (61%), Gaps = 1/118 (0%)
Frame = +1
Query: 2 LPNELVTNVSSNQTLANQWVQTNLVPFYSKTLIRYLLVGNELISSTTNQTWPHIVPAMYR 61
+PNEL+++ +++Q+ + WV+ N+ ++ T I + VGNE+ N T +V AM
Sbjct: 292 MPNELLSSAAADQSFTDAWVKANISSYHPATQIEAIAVGNEVFVDPKNTT-KFLVAAMKN 468
Query: 62 MKHSLTIFGLHK-VKVGTPLAMDVLQTSFPPSNGTFRNDIALSVMKPMLEFLHVTNSF 118
+ SLT GL +K+ +P+A+ LQ S+P S G+F+ ++ V+KPML+FL T S+
Sbjct: 469 VHASLTKQGLDSAIKISSPVALSALQNSYPASAGSFKTELIEPVIKPMLDFLRQTGSY 642
>TC14402 similar to UP|Q8L9D9 (Q8L9D9) Beta-1,3-glucanase-like protein,
partial (20%)
Length = 642
Score = 79.3 bits (194), Expect = 1e-15
Identities = 32/81 (39%), Positives = 50/81 (61%)
Frame = +1
Query: 291 GKAWCVVAEGANKTAVVEALSYACSQGNRTCELVQPGKPCFEPDSVVGHASYAFSSYWAQ 350
G+ WCV G ++ + AL YAC +G C +QPG C++P+++ HAS+AF+SY+ +
Sbjct: 49 GRTWCVANGGYSEEKLQAALDYACGEGGADCSAIQPGATCYDPNTLEAHASFAFNSYYQK 228
Query: 351 FRRVGGTCNFNGLATQIAEDP 371
R GTC+F G A + + P
Sbjct: 229 KARGTGTCDFGGAAYVVTQAP 291
>BP069771
Length = 529
Score = 75.9 bits (185), Expect = 1e-14
Identities = 41/109 (37%), Positives = 58/109 (52%)
Frame = -3
Query: 164 DQMVDAVYFAMERLGYPDIQIFIAETGWPNDGDLDQIGANIHNAGTYNRNFVKKVTKKPP 223
D MVDAV AM GY I I + ETGWP+ D+ AN+ A TY R V ++
Sbjct: 527 DMMVDAVASAMAEAGYETIPIVVTETGWPSSDAADEFNANLGYAETYLRGLVNHLSS--G 354
Query: 224 VGTPARPGSILPSFIFALYNENLKTGLGTERHFGLLYPNGSRIYEIDLS 272
GTP + +++ L++ + G T R +G+LYPNG+ Y +D S
Sbjct: 353 TGTPLLKDGVQQVYVYELFD---RKGTTTGRDWGILYPNGTAKYHVDFS 216
>AV412757
Length = 315
Score = 74.3 bits (181), Expect = 3e-14
Identities = 39/106 (36%), Positives = 63/106 (58%)
Frame = +2
Query: 91 PSNGTFRNDIALSVMKPMLEFLHVTNSFFFLDVYPFFAWTSDPININLDYALFESDNITV 150
PS+G FR DI ++M +++FL N+ F +++YPF + SD N DYA F +
Sbjct: 5 PSDGDFRPDIQ-NLMLQIVKFLSQNNAPFTVNIYPFISLYSDA-NFPGDYAFFNGFQSPI 178
Query: 151 TDSGTGLVYTNLFDQMVDAVYFAMERLGYPDIQIFIAETGWPNDGD 196
TD+G +Y N+ D D + +A+++ G+ ++ I + E GWP DGD
Sbjct: 179 TDNGK--IYDNVLDANYDTLVWALQKNGFGNLPIIVGEIGWPTDGD 310
>TC9275 similar to UP|Q9M4A9 (Q9M4A9) Beta-1,3 glucanase precursor ,
partial (39%)
Length = 545
Score = 72.4 bits (176), Expect = 1e-13
Identities = 37/96 (38%), Positives = 58/96 (59%)
Frame = +1
Query: 17 ANQWVQTNLVPFYSKTLIRYLLVGNELISSTTNQTWPHIVPAMYRMKHSLTIFGLHKVKV 76
A QWV T++ PF+ +T I Y+LVG+E++ +VPAM + +L G+ +KV
Sbjct: 256 ARQWVVTHIKPFHPQTRINYILVGSEVLHWGDTNMIRGLVPAMRTLYSALQAEGIRDIKV 435
Query: 77 GTPLAMDVLQTSFPPSNGTFRNDIALSVMKPMLEFL 112
T ++ ++++S PPS G FR A V+ PML+FL
Sbjct: 436 TTAHSLAIMRSSLPPSAGRFRPGFAKHVLGPMLKFL 543
>AV425443
Length = 372
Score = 71.6 bits (174), Expect = 2e-13
Identities = 39/105 (37%), Positives = 57/105 (54%), Gaps = 4/105 (3%)
Frame = +3
Query: 272 SGKTPEYEYKPLPP----PDDYKGKAWCVVAEGANKTAVVEALSYACSQGNRTCELVQPG 327
SG P +P P P +G++WCV GA + ++ AL YAC +G C +Q G
Sbjct: 54 SGSVPVINPQPPPANTNNPPSTQGQSWCVAKTGAPQASLQAALDYACGKG-ADCAALQQG 230
Query: 328 KPCFEPDSVVGHASYAFSSYWAQFRRVGGTCNFNGLATQIAEDPS 372
C+ P ++ HASYAF+SY+ Q +C+F G AT + +PS
Sbjct: 231 GSCYSPVTLQNHASYAFNSYY-QKNPAPTSCDFGGAATLVNTNPS 362
>AV768339
Length = 576
Score = 65.5 bits (158), Expect = 1e-11
Identities = 27/61 (44%), Positives = 40/61 (65%)
Frame = -3
Query: 312 YACSQGNRTCELVQPGKPCFEPDSVVGHASYAFSSYWAQFRRVGGTCNFNGLATQIAEDP 371
+AC G C + G+PC EPD+VV HA+YAF++Y+ + + GTC+F G+A + DP
Sbjct: 571 WACGPGKVDCSPLLQGQPCHEPDNVVAHATYAFNAYYQKMAKSPGTCDFKGVAAITSTDP 392
Query: 372 S 372
S
Sbjct: 391 S 389
>TC12695 similar to UP|Q9SAK1 (Q9SAK1) T8K14.10, partial (17%)
Length = 571
Score = 60.1 bits (144), Expect(2) = 1e-10
Identities = 27/52 (51%), Positives = 36/52 (68%)
Frame = +1
Query: 309 ALSYACSQGNRTCELVQPGKPCFEPDSVVGHASYAFSSYWAQFRRVGGTCNF 360
A+ YAC G C+LVQP PCF+P++V+ HASYAF+SY+ + GGT F
Sbjct: 1 AMDYACGSG-ADCKLVQPNGPCFQPNTVLAHASYAFNSYFQNTKNGGGTL*F 153
Score = 21.9 bits (45), Expect(2) = 1e-10
Identities = 9/21 (42%), Positives = 11/21 (51%)
Frame = +2
Query: 352 RRVGGTCNFNGLATQIAEDPS 372
R V C+F G A + DPS
Sbjct: 128 RMVEAPCDFGGTAMLVTVDPS 190
>TC12989 similar to GB|AAO39944.1|28372924|BT003716 At5g08000 {Arabidopsis
thaliana;}, partial (41%)
Length = 430
Score = 61.2 bits (147), Expect = 3e-10
Identities = 30/80 (37%), Positives = 47/80 (58%)
Frame = +2
Query: 293 AWCVVAEGANKTAVVEALSYACSQGNRTCELVQPGKPCFEPDSVVGHASYAFSSYWAQFR 352
+WCV +G N+ ++ +AL YAC G C ++ PC+ P++V H +YA +SY+ +
Sbjct: 188 SWCVCKDG-NEASLQKALDYACGAG-ADCNPIKQTGPCYNPNTVRAHCNYAVNSYFQKKG 361
Query: 353 RVGGTCNFNGLATQIAEDPS 372
+ C+F G AT A DPS
Sbjct: 362 QAPLACDFAGAATVSASDPS 421
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.321 0.138 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,854,306
Number of Sequences: 28460
Number of extensions: 97969
Number of successful extensions: 539
Number of sequences better than 10.0: 70
Number of HSP's better than 10.0 without gapping: 506
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 512
length of query: 389
length of database: 4,897,600
effective HSP length: 92
effective length of query: 297
effective length of database: 2,279,280
effective search space: 676946160
effective search space used: 676946160
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 56 (26.2 bits)
Medicago: description of AC136841.5


