

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136507.9 + phase: 0 /pseudo
(261 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
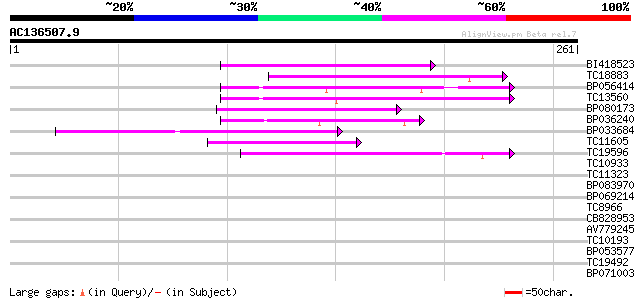
Score E
Sequences producing significant alignments: (bits) Value
BI418523 63 6e-11
TC18883 59 1e-09
BP056414 58 1e-09
TC13560 56 7e-09
BP080173 54 4e-08
BP036240 48 1e-06
BP033684 43 6e-05
TC11605 42 1e-04
TC19596 40 5e-04
TC10933 similar to UP|Q9LNS0 (Q9LNS0) F1L3.4, partial (6%) 39 0.001
TC11323 37 0.003
BP083970 34 0.029
BP069214 30 0.42
TC8966 similar to UP|Q9M5Q3 (Q9M5Q3) S-ribonuclease binding prot... 30 0.42
CB828953 29 0.72
AV779245 28 1.2
TC10193 similar to UP|Q944L7 (Q944L7) At1g18480/F15H18_1, partia... 28 1.2
BP053577 28 1.6
TC19492 similar to UP|O22792 (O22792) At2g33420 protein, partial... 28 1.6
BP071003 28 2.1
>BI418523
Length = 496
Score = 62.8 bits (151), Expect = 6e-11
Identities = 35/99 (35%), Positives = 50/99 (50%)
Frame = +1
Query: 98 WIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNLNTNSAFNAKLLGVIT 157
W PP +IK N DGA NP + GGVFRD N +G F N A+ AK+ ++
Sbjct: 187 WSPPSAGFIKCNVDGASQGNPGPSGVGGVFRDANRKILGYFSLNSGNGWAYEAKVRSILN 366
Query: 158 AINIAGENNWRNIWLETDLQLAVLAFKNLNMVPWSLRNK 196
A+ + +NI +E+D +AV + PW L N+
Sbjct: 367 ALVFIQKFLLKNILIESDSTVAVEWVMSKANRPWRLSNE 483
>TC18883
Length = 821
Score = 58.5 bits (140), Expect = 1e-09
Identities = 35/111 (31%), Positives = 55/111 (49%), Gaps = 1/111 (0%)
Frame = +1
Query: 120 KASCGGVFRDHNNIFIGAFVQNLNTNSAFNAKLLGVITAINIAGENNWRNIWLETDLQLA 179
+ GG+FR+ +N +G F +++ AF A++ + A+ I RN+ +E+D LA
Sbjct: 4 RCGIGGIFRNSDNEILGFFSKHVGEGFAFEAEVWAIFEALTICVNRQLRNLTIESDSSLA 183
Query: 180 VLAFKNLNMVPWSLRNKWLNYFEHVRSMNFL-ITHIYIEGNSCADCMANVG 229
V N + PW L NK V +N L + HI+ E N D +A G
Sbjct: 184 VGWVNNRSXRPWKLLNKLNQIDLWVAEVNCLSVKHIFREANGEVDKLAKRG 336
>BP056414
Length = 606
Score = 58.2 bits (139), Expect = 1e-09
Identities = 45/146 (30%), Positives = 68/146 (45%), Gaps = 11/146 (7%)
Frame = -3
Query: 98 WIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNLNT---NSAFNAKLLG 154
W P W KVN DGAL+ +ASCGGV RD + F ++L + AF +L+
Sbjct: 562 WTRPPEGWFKVNVDGALS-GDLQASCGGVVRDAAGSWSKGFARSLGVLRWHXAFFVELMA 386
Query: 155 VITAINIAGENNWRNIWLETDLQLAVLAFKNLNM--------VPWSLRNKWLNYFEHVRS 206
V TA++ + + +E+D Q V +N N + W ++ K +
Sbjct: 385 VQTAVDFIMSWDIPQVIIESDSQQVVELLQNFNASSPHQYGEIAWDIQQKISVH------ 224
Query: 207 MNFLITHIYIEGNSCADCMANVGLTL 232
+ +IT E N AD +A VGL+L
Sbjct: 223 GSIVITSTPREANFLADYLAKVGLSL 146
>TC13560
Length = 571
Score = 55.8 bits (133), Expect = 7e-09
Identities = 40/138 (28%), Positives = 65/138 (46%), Gaps = 3/138 (2%)
Frame = +2
Query: 98 WIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNLNTNSAFN---AKLLG 154
WI P W+K+N DGA++ N +ASC GV R+ + ++ F N T S+ N +L+
Sbjct: 146 WIKPDAGWVKINVDGAVS-NCLQASCRGVLRNADGSWLKGFRWNFGTFSSANVFLTELMA 322
Query: 155 VITAINIAGENNWRNIWLETDLQLAVLAFKNLNMVPWSLRNKWLNYFEHVRSMNFLITHI 214
V TA+ A + + +E+D V + ++ L+ R L+
Sbjct: 323 VKTAVEAAMSLDLARVIVESDSLEVVDFLHSTDLGSHRYSQIALDILHLQREHGALVFQY 502
Query: 215 YIEGNSCADCMANVGLTL 232
+ GNS D +A GL+L
Sbjct: 503 ALRGNSIVDYIAKFGLSL 556
>BP080173
Length = 382
Score = 53.5 bits (127), Expect = 4e-08
Identities = 28/85 (32%), Positives = 40/85 (46%)
Frame = -2
Query: 96 VMWIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNLNTNSAFNAKLLGV 155
V W P+ WIK+N DG+ + A+CGGV DH FI L +A+L G+
Sbjct: 351 VRWPVPMEGWIKINIDGSFAPSSNCAACGGVMWDHEGTFITGASVRLGCYFINHAELWGI 172
Query: 156 ITAINIAGENNWRNIWLETDLQLAV 180
IA ++ I +E+D V
Sbjct: 171 FHGAKIAQARGFKKILVESDSSFVV 97
>BP036240
Length = 567
Score = 48.1 bits (113), Expect = 1e-06
Identities = 31/99 (31%), Positives = 55/99 (55%), Gaps = 5/99 (5%)
Frame = -3
Query: 98 WIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQN---LNTNSAFNAKLLG 154
W P W K + DGAL+++ ASCGGV RD +I F +N L ++ F +L
Sbjct: 562 WTAPDEGWTKFDVDGALSRD-LLASCGGVLRDSRRHWIKGFCRNMGDLTYHNVFLVELSA 386
Query: 155 VITAINIAGENNWRNIWLETDLQLAV--LAFKNLNMVPW 191
+ +A+ IA + +++ +E+D A+ L+ +N++ P+
Sbjct: 385 IQSAVEIALSMDLQHVIVESDALEAIHLLSSENVHAHPF 269
>BP033684
Length = 557
Score = 42.7 bits (99), Expect = 6e-05
Identities = 32/132 (24%), Positives = 57/132 (42%)
Frame = -1
Query: 22 VIINILSAIWFIRNQARFKDKKNHWKSAIDNIIVAVSLTGNRTKLTSFVNMDEFVFLKAF 81
++ ++ ++W R Q+ FK K + D ++ + N D+F + F
Sbjct: 509 ILYALVWSLWLERKQSVFKGKLASSQEVWDLHLMTIFWWIKAVWKECPYNFDQFH--RDF 336
Query: 82 KINIHPPRAPLIKEVMWIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQN 141
+ P + W PP +K N DGA NP + GGV D+ ++ +G F N
Sbjct: 335 CMLSFKKLIPKQRLKGWTPPSRGILKFNVDGASQGNPGPSGVGGVLCDYRSMVLGFFSIN 156
Query: 142 LNTNSAFNAKLL 153
+ A+ A++L
Sbjct: 155 MGHGWAYEAEVL 120
>TC11605
Length = 546
Score = 42.0 bits (97), Expect = 1e-04
Identities = 17/71 (23%), Positives = 37/71 (51%)
Frame = +3
Query: 92 LIKEVMWIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNLNTNSAFNAK 151
+++ +W PP +K N DG+ +P + GG+ R+ + +G F + L A+ A+
Sbjct: 330 MVRNCVWCPPEANILKFNVDGSARGSPGISGAGGILRNAESAVLGRFSKPLGVLCAYQAE 509
Query: 152 LLGVITAINIA 162
+ ++ A+ +
Sbjct: 510 VKAILLALKFS 542
>TC19596
Length = 499
Score = 39.7 bits (91), Expect = 5e-04
Identities = 29/128 (22%), Positives = 58/128 (44%), Gaps = 2/128 (1%)
Frame = +1
Query: 107 KVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNLNTNSAFNAKLLGVITAINIAGENN 166
++++DGA + + GG+ RD + +I F A A++ + + + +
Sbjct: 4 RMDSDGAFKHDDDRMGMGGIVRDAHGAWISGFYAGSLGGDALRAEIAALKHGLTLLWNAH 183
Query: 167 WRNIWLETDLQLAVLAFKNLNMVPWSLRNKWLNYFEHVRSMNFLITHIYI--EGNSCADC 224
R E D V A +N +L ++ L+ + ++ +T Y+ E N+ ADC
Sbjct: 184 VRRATCEVDCLDIVEALENDRYQFHALASELLD-IRLLLDRDWTVTLAYVPREANAAADC 360
Query: 225 MANVGLTL 232
+A +G +L
Sbjct: 361 LAGLGASL 384
>TC10933 similar to UP|Q9LNS0 (Q9LNS0) F1L3.4, partial (6%)
Length = 596
Score = 38.5 bits (88), Expect = 0.001
Identities = 13/29 (44%), Positives = 19/29 (64%)
Frame = +2
Query: 98 WIPPIPPWIKVNTDGALTKNPTKASCGGV 126
W PP W+K+NTDG+ T+ + CGG+
Sbjct: 107 WTPPSVGWVKLNTDGSFTRGSSLMGCGGL 193
>TC11323
Length = 657
Score = 37.4 bits (85), Expect = 0.003
Identities = 23/96 (23%), Positives = 40/96 (40%)
Frame = +2
Query: 98 WIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNLNTNSAFNAKLLGVIT 157
W+ P+ K+N DG+ N A GG+ RD ++ FI F L L ++
Sbjct: 5 WLKPVHGRFKLNFDGSSLGNRGNAGGGGLLRDGSSNFIFGFSIFLAVAQIMKLSLCAILE 184
Query: 158 AINIAGENNWRNIWLETDLQLAVLAFKNLNMVPWSL 193
+ + + I +E D + V ++ + W L
Sbjct: 185 GLLVCKPLGYDGIGIECDSNIVVSWIRSRSCQLWYL 292
>BP083970
Length = 375
Score = 33.9 bits (76), Expect = 0.029
Identities = 26/64 (40%), Positives = 36/64 (55%), Gaps = 2/64 (3%)
Frame = -3
Query: 168 RNIWLETDLQLAVLAFKNLNMVPWSLRNKWLNYFEH-VRSMNFL-ITHIYIEGNSCADCM 225
RNI + +D LAV N + PW L + LN +H + +N L ++HIY E NS AD +
Sbjct: 367 RNISI*SDSSLAVGWINNCSNRPWKLLS-MLNPIDHWLVEVNCLKVSHIYREANSEADKL 191
Query: 226 ANVG 229
A G
Sbjct: 190 AKRG 179
>BP069214
Length = 545
Score = 30.0 bits (66), Expect = 0.42
Identities = 15/38 (39%), Positives = 19/38 (49%)
Frame = -2
Query: 205 RSMNFLITHIYIEGNSCADCMANVGLTLSSFVWLPSLP 242
R +TH+ EGNSCAD +A G + LP P
Sbjct: 229 REWEVSMTHMLREGNSCADFLAKYGAMQDEPLVLPQEP 116
>TC8966 similar to UP|Q9M5Q3 (Q9M5Q3) S-ribonuclease binding protein SBP1,
partial (48%)
Length = 611
Score = 30.0 bits (66), Expect = 0.42
Identities = 12/37 (32%), Positives = 23/37 (61%), Gaps = 1/37 (2%)
Frame = +3
Query: 176 LQLAVLAFKN-LNMVPWSLRNKWLNYFEHVRSMNFLI 211
+ LA + + N LN+ W W+ YF+H++S+ F++
Sbjct: 387 VSLAFVLYANPLNLSAWRSSCNWIFYFDHLKSLIFIL 497
>CB828953
Length = 554
Score = 29.3 bits (64), Expect = 0.72
Identities = 13/50 (26%), Positives = 22/50 (44%)
Frame = -3
Query: 98 WIPPIPPWIKVNTDGALTKNPTKASCGGVFRDHNNIFIGAFVQNLNTNSA 147
W PP+P +K+N D + + S G RD + + ++SA
Sbjct: 540 WRPPLPRMLKLNVDASWKASSPLCSIGIAVRDEAGLLVAGSASKAFSHSA 391
>AV779245
Length = 487
Score = 28.5 bits (62), Expect = 1.2
Identities = 9/26 (34%), Positives = 15/26 (57%)
Frame = -2
Query: 217 EGNSCADCMANVGLTLSSFVWLPSLP 242
+GN D +A + +L FVW+ +P
Sbjct: 255 QGNRAVDALATLSFSLGDFVWIEEIP 178
>TC10193 similar to UP|Q944L7 (Q944L7) At1g18480/F15H18_1, partial (22%)
Length = 793
Score = 28.5 bits (62), Expect = 1.2
Identities = 20/66 (30%), Positives = 29/66 (43%), Gaps = 4/66 (6%)
Frame = -1
Query: 44 NHWKSAIDNIIVAVSLTGNRTKLTSFVNMDEFVFL----KAFKINIHPPRAPLIKEVMWI 99
NH K +N A++ T ++ N F F K F++NI P PLI +
Sbjct: 352 NHEKHRAENSH*ALTSTCLGISISGTNNPISFSFFAF*YKGFEVNIRKPEFPLISKTSGK 173
Query: 100 PPIPPW 105
PP P+
Sbjct: 172 PPPHPF 155
>BP053577
Length = 463
Score = 28.1 bits (61), Expect = 1.6
Identities = 31/133 (23%), Positives = 58/133 (43%), Gaps = 11/133 (8%)
Frame = +3
Query: 121 ASCGGVFRD-HNNIFIGAFVQNLNTNSAFNAKLLGVITAINIAGENNWRNIWLETD-LQL 178
A G V R+ + A ++ +S A+ LG+ + +A + +R I LETD LQL
Sbjct: 12 AGLGMVARNCDGEVMASACSSPVSISSPLLAEALGLRWTMQLATDLGFRRITLETDCLQL 191
Query: 179 AVLAFKNLNMVPWSLRNKWLNYFEHVRS--------MNFL-ITHIYIEGNSCADCMANVG 229
W + + +Y + S +++ ++ + GN+ AD +A
Sbjct: 192 ---------FNTWRSQEEGRSYLSSIISDCRILISAFDYVSVSFVRRTGNTVADFLARNS 344
Query: 230 LTLSSFVWLPSLP 242
T ++ VW+ +P
Sbjct: 345 DTYANMVWVEEVP 383
>TC19492 similar to UP|O22792 (O22792) At2g33420 protein, partial (11%)
Length = 510
Score = 28.1 bits (61), Expect = 1.6
Identities = 17/42 (40%), Positives = 25/42 (59%), Gaps = 1/42 (2%)
Frame = +1
Query: 98 WIPPIPPWIKVNTDGALTKNPTKAS-CGGVFRDHNNIFIGAF 138
+IP +PP + N D TK KA+ CG F+D +++ GAF
Sbjct: 232 YIPMLPPLTRCNRDSKFTKLWKKAAPCGANFQDLHHM-KGAF 354
>BP071003
Length = 466
Score = 27.7 bits (60), Expect = 2.1
Identities = 11/28 (39%), Positives = 17/28 (60%)
Frame = +3
Query: 205 RSMNFLITHIYIEGNSCADCMANVGLTL 232
R+ ITH+ EGN DC+A +G ++
Sbjct: 132 RAWEVSITHVPREGNEPTDCLAGLGASM 215
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.325 0.139 0.455
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,716,949
Number of Sequences: 28460
Number of extensions: 86881
Number of successful extensions: 626
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 623
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 624
length of query: 261
length of database: 4,897,600
effective HSP length: 88
effective length of query: 173
effective length of database: 2,393,120
effective search space: 414009760
effective search space used: 414009760
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 54 (25.4 bits)
Medicago: description of AC136507.9


