

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136507.3 + phase: 0 /pseudo
(491 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
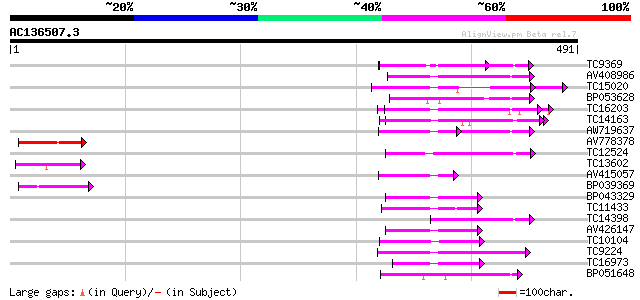
Score E
Sequences producing significant alignments: (bits) Value
TC9369 similar to UP|Q93X72 (Q93X72) Leucine-rich repeat resista... 58 3e-09
AV408986 58 4e-09
TC15020 homologue to UP|Q9QX99 (Q9QX99) Transcription factor (T-... 57 7e-09
BP053628 57 7e-09
TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodula... 57 9e-09
TC14163 57 9e-09
AW719637 55 2e-08
AV778378 55 2e-08
TC12524 similar to UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kina... 54 6e-08
TC13602 weakly similar to UP|Q84QD8 (Q84QD8) Avr9/Cf-9 rapidly e... 52 3e-07
AV415057 49 1e-06
BP039369 49 2e-06
BP043329 49 2e-06
TC11433 similar to UP|AAR99871 (AAR99871) Strubbelig receptor fa... 47 5e-06
TC14398 similar to UP|Q96477 (Q96477) LRR protein, partial (86%) 47 7e-06
AV426147 46 2e-05
TC10104 similar to UP|Q8GTD6 (Q8GTD6) Polygalacturonase inhibito... 44 5e-05
TC9224 similar to GB|AAP21230.1|30102624|BT006422 At3g49750 {Ara... 44 5e-05
TC16973 weakly similar to UP|Q96477 (Q96477) LRR protein, partia... 44 6e-05
BP051648 42 2e-04
>TC9369 similar to UP|Q93X72 (Q93X72) Leucine-rich repeat resistance
protein-like protein, partial (70%)
Length = 859
Score = 58.2 bits (139), Expect = 3e-09
Identities = 44/134 (32%), Positives = 67/134 (49%), Gaps = 1/134 (0%)
Frame = +1
Query: 321 LIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASN 380
L + N L G IP G+L +LR LD +N L + ++ GC +L+ LYL +N
Sbjct: 238 LYLHENRLIGRIPPELGTLQNLRHLDAGNNHLVGTIRELIR--IEGCFP-ALRNLYLNNN 408
Query: 381 QIIGTVP-DMSGFSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVITDSHFG 439
G +P ++ SSLE ++L N ++G I + +L LYLD N G I + F
Sbjct: 409 YFTGGMPAQLANLSSLEILYLSYNKMSGVIPSSLAHIPKLTYLYLDHNQFSGRIPEP-FY 585
Query: 440 NMSMLKYLSLSSNS 453
S LK + + N+
Sbjct: 586 KHSFLKEMYIEGNA 627
Score = 40.4 bits (93), Expect = 7e-04
Identities = 29/97 (29%), Positives = 44/97 (44%)
Frame = +1
Query: 320 SLIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLAS 379
+L + +N GG+P +L SL L LS NK+S +P L ++ L LYL
Sbjct: 388 NLYLNNNYFTGGMPAQLANLSSLEILYLSYNKMSGVIPSSLAHIP------KLTYLYLDH 549
Query: 380 NQIIGTVPDMSGFSSLENMFLYENLLNGTILKNSTFP 416
NQ G +P+ ++ FL E + G + P
Sbjct: 550 NQFSGRIPE----PFYKHSFLKEMYIEGNAFRPGVKP 648
Score = 38.5 bits (88), Expect = 0.003
Identities = 35/114 (30%), Positives = 51/114 (44%), Gaps = 1/114 (0%)
Frame = +1
Query: 342 LRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASNQIIGTV-PDMSGFSSLENMFL 400
L LDL +NKL+ +P + L LK L L N++ + P++ SL +++L
Sbjct: 13 LTRLDLHNNKLTGPIPPQIGRL------KRLKILNLRWNKLQDAIPPEIGELKSLTHLYL 174
Query: 401 YENLLNGTILKNSTFPYRLANLYLDSNDLDGVITDSHFGNMSMLKYLSLSSNSL 454
N G I K L LYL N L G I G + L++L +N L
Sbjct: 175 SFNSFKGEIPKELANLPDLRYLYLHENRLIGRI-PPELGTLQNLRHLDAGNNHL 333
>AV408986
Length = 434
Score = 57.8 bits (138), Expect = 4e-09
Identities = 46/128 (35%), Positives = 64/128 (49%), Gaps = 1/128 (0%)
Frame = +3
Query: 328 LEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASNQIIGTVP 387
L G IP G++ SL L L N S +P L LS LK LY+ +NQ+ GT+P
Sbjct: 3 LSGEIPPEIGNISSLELLALHQNSFSGAIPKELGKLS------GLKRLYVYTNQLNGTIP 164
Query: 388 -DMSGFSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVITDSHFGNMSMLKY 446
++ ++ + L EN L G I K L+ L+L N+L G I G++ LK
Sbjct: 165 TELGNCTNAIEIDLSENRLIGIIPKELGQISNLSLLHLFENNLQGHI-PRELGSLRQLKK 341
Query: 447 LSLSSNSL 454
L LS N+L
Sbjct: 342 LDLSLNNL 365
Score = 39.3 bits (90), Expect = 0.001
Identities = 18/39 (46%), Positives = 26/39 (66%)
Frame = +3
Query: 326 NSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLS 364
N+L+G IP+ GSL L+ LDLS N L+ +P+ NL+
Sbjct: 285 NNLQGHIPRELGSLRQLKKLDLSLNNLTGTIPLEFQNLT 401
>TC15020 homologue to UP|Q9QX99 (Q9QX99) Transcription factor (T-cell
leukemia, homeobox 1), partial (5%)
Length = 1304
Score = 57.0 bits (136), Expect = 7e-09
Identities = 53/173 (30%), Positives = 81/173 (46%), Gaps = 3/173 (1%)
Frame = +1
Query: 314 SVQS*HSLIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLK 373
+++S +SL NSL G +P S SL +LR +DLS NKL+ +P + N L
Sbjct: 439 TLKSLYSLDFSHNSLSGSLPNSMNSLINLRRIDLSFNKLTGSIPKLPPN---------LL 591
Query: 374 ELYLASNQIIGTV--PDMSGFSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDG 431
EL + +N + G + G + LE + L EN L GT+ L + L +N G
Sbjct: 592 ELAVKANSLSGPLQKATFEGLNQLEVVELSENTLAGTVEPWFFLLPSLQQVDLSNNSFTG 771
Query: 432 V-ITDSHFGNMSMLKYLSLSSNSLALKFSENWVPPFQLSTIYLRSCTLGPSFP 483
V I+ G S L ++L N + N QLS++ +R +L + P
Sbjct: 772 VQISRPARGEGSNLVAVNLGFNRIQGYAPANLAAYPQLSSLSIRHNSLRGAIP 930
Score = 54.7 bits (130), Expect = 3e-08
Identities = 40/142 (28%), Positives = 67/142 (47%)
Frame = +1
Query: 314 SVQS*HSLIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLK 373
S+ + +L ++SNS G IP S +L SL SLD S N LS LP +++L +L+
Sbjct: 367 SLSNLKTLTLRSNSFSGSIPPSITTLKSLYSLDFSHNSLSGSLPNSMNSLI------NLR 528
Query: 374 ELYLASNQIIGTVPDMSGFSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVI 433
+ L+ N++ G++P + P L L + +N L G +
Sbjct: 529 RIDLSFNKLTGSIPKL--------------------------PPNLLELAVKANSLSGPL 630
Query: 434 TDSHFGNMSMLKYLSLSSNSLA 455
+ F ++ L+ + LS N+LA
Sbjct: 631 QKATFEGLNQLEVVELSENTLA 696
>BP053628
Length = 567
Score = 57.0 bits (136), Expect = 7e-09
Identities = 48/167 (28%), Positives = 72/167 (42%), Gaps = 42/167 (25%)
Frame = +1
Query: 330 GGIPKSFGSLCSLRSLDLSSNKLSEDLPVML---------------------------HN 362
G +P S L +L +LDLSSN L +P + N
Sbjct: 31 GSLPSSIHRLYALEALDLSSNYLYGSIPPKIFTIGNLQTLRLGDNFLNGTIPTLFNSSSN 210
Query: 363 LSVGCAKN---------------SLKELYLASNQIIGTVPDMSGFSSLENMFLYENLLNG 407
L V KN +L E+ +++N+I G++ D +G SLE + L EN L+
Sbjct: 211 LKVLSLKNNRLTGQFPSSILSITTLIEIDMSTNEISGSLQDFTGLRSLEQLDLRENRLDS 390
Query: 408 TILKNSTFPYRLANLYLDSNDLDGVITDSHFGNMSMLKYLSLSSNSL 454
+ K P RL ++L N G I H+G + ML+ L +S N+L
Sbjct: 391 ALPK---MPPRLMGIFLSRNSFSGQI-PKHYGQLKMLQQLDVSFNAL 519
>TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodulation
aberrant root formation protein), complete
Length = 3308
Score = 56.6 bits (135), Expect = 9e-09
Identities = 41/138 (29%), Positives = 72/138 (51%), Gaps = 1/138 (0%)
Frame = +2
Query: 325 SNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASNQIIG 384
SN+ EGGIP +FGS+ +LR L++++ L+ ++P L NL+ L L++ N + G
Sbjct: 794 SNAYEGGIPPAFGSMENLRLLEMANCNLTGEIPPSLGNLT------KLHSLFVQMNNLTG 955
Query: 385 TV-PDMSGFSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVITDSHFGNMSM 443
T+ P++S SL ++ L N L G I ++ + L + N G + S G++
Sbjct: 956 TIPPELSSMMSLMSLDLSINDLTGEIPESFSKLKNLTLMNFFQNKFRGSL-PSFIGDLPN 1132
Query: 444 LKYLSLSSNSLALKFSEN 461
L+ L + N+ + N
Sbjct: 1133LETLQVWENNFSFVLPHN 1186
Score = 49.3 bits (116), Expect = 1e-06
Identities = 48/174 (27%), Positives = 76/174 (43%), Gaps = 21/174 (12%)
Frame = +2
Query: 319 HSLIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLA 378
HSL ++ N+L G IP S+ SL SLDLS N L+ ++P L +L +
Sbjct: 920 HSLFVQMNNLTGTIPPELSSMMSLMSLDLSINDLTGEIPESFSKL------KNLTLMNFF 1081
Query: 379 SNQIIGTVPDMSG-FSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDG------ 431
N+ G++P G +LE + ++EN + + N R + N L G
Sbjct: 1082 QNKFRGSLPSFIGDLPNLETLQVWENNFSFVLPHNLGGNGRFLYFDVTKNHLTGLIPPDL 1261
Query: 432 ---------VITDSHFGN---MSMLKYLSLSSNSLALKFSENWVPP--FQLSTI 471
+ITD+ F + + SL+ +A F + VPP FQL ++
Sbjct: 1262 CKSGRLKTFIITDNFFRGPIPKGIGECRSLTKIRVANNFLDGPVPPGVFQLPSV 1423
Score = 28.5 bits (62), Expect = 2.6
Identities = 13/37 (35%), Positives = 20/37 (53%)
Frame = +2
Query: 321 LIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLP 357
L + N + G +P + SL +LDLSSN + +P
Sbjct: 1787 LNLSRNEISGPVPDEIRFMTSLTTLDLSSNNFTGTVP 1897
>TC14163
Length = 1712
Score = 56.6 bits (135), Expect = 9e-09
Identities = 44/144 (30%), Positives = 66/144 (45%), Gaps = 1/144 (0%)
Frame = +1
Query: 321 LIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASN 380
L ++ N G IP F S L L LS NK S +P + L+ L+ L L N
Sbjct: 718 LSLEGNQFSGAIPDFFSSFTDLGILRLSRNKFSGKIPASISTLAP-----KLRYLELGHN 882
Query: 381 QIIGTVPDMSG-FSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVITDSHFG 439
Q+ G +PD G F +L+ + L N +GT+ + ++ NL L +N L +
Sbjct: 883 QLSGKIPDFLGKFRALDTLDLSSNRFSGTVPASFKNLTKIFNLNLANNLLVDPFPEM--- 1053
Query: 440 NMSMLKYLSLSSNSLALKFSENWV 463
N+ ++ L LS+N L WV
Sbjct: 1054NVKGIESLDLSNNMFHLNQIPKWV 1125
Score = 43.5 bits (101), Expect = 8e-05
Identities = 39/146 (26%), Positives = 64/146 (43%), Gaps = 5/146 (3%)
Frame = +1
Query: 326 NSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASNQIIGT 385
N L G IP G +L +LDLSSN+ S +P NL+ + L LA+N ++
Sbjct: 880 NQLSGKIPDFLGKFRALDTLDLSSNRFSGTVPASFKNLT------KIFNLNLANNLLVDP 1041
Query: 386 VPDMS--GFSSLE---NMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVITDSHFGN 440
P+M+ G SL+ NMF + F +LA + D +++F +
Sbjct: 1042 FPEMNVKGIESLDLSNNMFHLNQIPKWVTSSPIIFSLKLARCGIKMKLDDWKPAETYFYD 1221
Query: 441 MSMLKYLSLSSNSLALKFSENWVPPF 466
L +S +++ L S ++ F
Sbjct: 1222 FIDLSGNEISGSAVGLVNSTEYLVGF 1299
Score = 37.0 bits (84), Expect = 0.007
Identities = 19/61 (31%), Positives = 28/61 (45%)
Frame = +1
Query: 8 CIEKERHALLELKSGLVLDDTYLLPSWDTKSDDCCAWEGIGCRNQTGHVEILDLNSDQFG 67
C + L+ KSG+ D + +L SW DCC W+G+ C V L L+ +
Sbjct: 187 CNVDDEAGLMGFKSGIKSDPSGILKSW-IPGTDCCTWQGVTCLFDDKRVTSLYLSGNPEN 363
Query: 68 P 68
P
Sbjct: 364 P 366
>AW719637
Length = 571
Score = 55.5 bits (132), Expect = 2e-08
Identities = 46/137 (33%), Positives = 68/137 (49%), Gaps = 2/137 (1%)
Frame = +2
Query: 320 SLIIKSNSLEGGIPKSFGSLCSLRSLDLSSNK-LSEDLPVMLHNLSVGCAKNSLKELYLA 378
+L + N L G IPKS GSL +L+ NK L ++P + N + SL L LA
Sbjct: 122 NLTLFDNHLSGEIPKSIGSLRNLQVFRAGGNKNLKGEIPWEIGNCT------SLVMLGLA 283
Query: 379 SNQIIGTVP-DMSGFSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVITDSH 437
I G++P + ++ + +Y LL+G+I + L NLYL N + G I S
Sbjct: 284 ETSISGSLPSSIQLLKRIKTIAIYTTLLSGSIPEEIGNCSELQNLYLYQNSISGSI-PSQ 460
Query: 438 FGNMSMLKYLSLSSNSL 454
G +S LK L L N++
Sbjct: 461 IGELSKLKSLLLWQNNI 511
Score = 42.4 bits (98), Expect = 2e-04
Identities = 24/72 (33%), Positives = 37/72 (51%)
Frame = +2
Query: 320 SLIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLAS 379
++ I + L G IP+ G+ L++L L N +S +P + LS LK L L
Sbjct: 341 TIAIYTTLLSGSIPEEIGNCSELQNLYLYQNSISGSIPSQIGELS------KLKSLLLWQ 502
Query: 380 NQIIGTVPDMSG 391
N I+GT+P+ G
Sbjct: 503 NNIVGTIPEEIG 538
>AV778378
Length = 555
Score = 55.5 bits (132), Expect = 2e-08
Identities = 29/59 (49%), Positives = 37/59 (62%)
Frame = +3
Query: 8 CIEKERHALLELKSGLVLDDTYLLPSWDTKSDDCCAWEGIGCRNQTGHVEILDLNSDQF 66
CI +ER ALLE K+ L + + L SW ++ C WEGIGC N TGHV LDL++ F
Sbjct: 174 CIGRERQALLEFKASLQKNPSNRLSSWKGTTNGC-EWEGIGCDNITGHVVKLDLSNPCF 347
>TC12524 similar to UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1,
partial (27%)
Length = 834
Score = 53.9 bits (128), Expect = 6e-08
Identities = 42/132 (31%), Positives = 67/132 (49%), Gaps = 2/132 (1%)
Frame = +2
Query: 326 NSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASNQIIGT 385
N+ G IP+S G L +DLSSNKL+ LP + C+ N L+ L N + G
Sbjct: 383 NNFTGSIPQSLGKNGKLTLVDLSSNKLTGTLPPHM------CSGNRLQTLIALGNFLFGP 544
Query: 386 VPDMSG-FSSLENMFLYENLLNGTILKNSTFPYRLANLYLDSNDLDGVITDSHFGNMS-M 443
+P+ G SL + + +N LNG+I K +L + N L G ++ G++S
Sbjct: 545 IPESLGKCESLTRIRMGQNFLNGSIPKGLFGLPKLTQVEFQDNLLSGEFPET--GSVSHN 718
Query: 444 LKYLSLSSNSLA 455
+ ++LS+N L+
Sbjct: 719 IGQITLSNNKLS 754
>TC13602 weakly similar to UP|Q84QD8 (Q84QD8) Avr9/Cf-9 rapidly elicited
protein 275 (Fragment), partial (26%)
Length = 556
Score = 51.6 bits (122), Expect = 3e-07
Identities = 25/68 (36%), Positives = 35/68 (50%), Gaps = 8/68 (11%)
Frame = +3
Query: 6 GSCIEKERHALLELKSGLVLDDTYL--------LPSWDTKSDDCCAWEGIGCRNQTGHVE 57
G C E + HALL+ K G + + SW+ +D C +W+GI C TGHV
Sbjct: 108 GQCHEDDSHALLQFKEGFAISKLASENPLSYPKVASWNASTDCCSSWDGIQCDEHTGHVI 287
Query: 58 ILDLNSDQ 65
+DL+S Q
Sbjct: 288 GIDLSSSQ 311
>AV415057
Length = 406
Score = 49.3 bits (116), Expect = 1e-06
Identities = 26/69 (37%), Positives = 39/69 (55%)
Frame = +3
Query: 320 SLIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLAS 379
+L +K+N L G IP G L L LDL +N S ++PV + NL+ +L+ L L+
Sbjct: 165 ALYLKNNRLSGSIPIEIGQLSVLHQLDLKNNNFSGNIPVQISNLT------NLETLDLSG 326
Query: 380 NQIIGTVPD 388
N + G +PD
Sbjct: 327 NHLSGEIPD 353
>BP039369
Length = 561
Score = 48.9 bits (115), Expect = 2e-06
Identities = 25/65 (38%), Positives = 34/65 (51%)
Frame = +2
Query: 8 CIEKERHALLELKSGLVLDDTYLLPSWDTKSDDCCAWEGIGCRNQTGHVEILDLNSDQFG 67
C+ R AL++ K+GL D L SW + +CC W GI C N TG V +DL +
Sbjct: 290 CLASNREALIDFKNGLE-DPFNRLSSWRNWNTNCCQWHGIHCDNITGAVVAIDLGNPHPH 466
Query: 68 PFEGD 72
P G+
Sbjct: 467 PVFGE 481
>BP043329
Length = 484
Score = 48.5 bits (114), Expect = 2e-06
Identities = 30/84 (35%), Positives = 44/84 (51%)
Frame = +3
Query: 326 NSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASNQIIGT 385
N L G IP S L SLR ++L +N LS +LP + NL+ L+ L + N + G
Sbjct: 207 NDLYGSIPSSLTGLTSLRQIELYNNSLSGELPRGMGNLT------ELRLLDASMNHLTGR 368
Query: 386 VPDMSGFSSLENMFLYENLLNGTI 409
+P+ LE++ LYEN G +
Sbjct: 369 IPEELCSLPLESLNLYENRFEGEL 440
>TC11433 similar to UP|AAR99871 (AAR99871) Strubbelig receptor family 3,
partial (24%)
Length = 565
Score = 47.4 bits (111), Expect = 5e-06
Identities = 33/87 (37%), Positives = 48/87 (54%)
Frame = +2
Query: 323 IKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASNQI 382
+ N L G IP +F SL L +LDLS+N LS +LP + NLS +L L L +NQ+
Sbjct: 272 LNENHLSGEIPDAFQSLTQLINLDLSTNNLSGELPPSVENLS------ALTTLRLQNNQL 433
Query: 383 IGTVPDMSGFSSLENMFLYENLLNGTI 409
GT+ D+ L+++ + N G I
Sbjct: 434 SGTL-DVLQDLPLQDLNVENNQFAGPI 511
>TC14398 similar to UP|Q96477 (Q96477) LRR protein, partial (86%)
Length = 982
Score = 47.0 bits (110), Expect = 7e-06
Identities = 32/91 (35%), Positives = 49/91 (53%), Gaps = 1/91 (1%)
Frame = +1
Query: 365 VGCAKNSLKELYLASNQIIG-TVPDMSGFSSLENMFLYENLLNGTILKNSTFPYRLANLY 423
V C NS+ + L + + G VPD+ SL+ + LYEN + GTI + L +L
Sbjct: 307 VTCQDNSVTRVDLGNLNLSGHLVPDLGNLHSLQYLELYENNIQGTIPEELGNLQSLISLD 486
Query: 424 LDSNDLDGVITDSHFGNMSMLKYLSLSSNSL 454
L N++ G I S GN+ L++L L++N L
Sbjct: 487 LYHNNVSGSIPSS-LGNLKNLRFLRLNNNHL 576
Score = 26.9 bits (58), Expect = 7.6
Identities = 19/63 (30%), Positives = 27/63 (42%), Gaps = 1/63 (1%)
Frame = +1
Query: 1 MLLVSGSCIEKERHALLELKSGLVLDDTYLLPSWDTKSDDCCAWEGIGCR-NQTGHVEIL 59
+ LV + E AL K L D +L SWD C W + C+ N V++
Sbjct: 172 LTLVPFAVANSEGDALYAFKQSLS-DPDNVLQSWDATLVSPCTWFHVTCQDNSVTRVDLG 348
Query: 60 DLN 62
+LN
Sbjct: 349 NLN 357
>AV426147
Length = 304
Score = 45.8 bits (107), Expect = 2e-05
Identities = 30/85 (35%), Positives = 42/85 (49%), Gaps = 1/85 (1%)
Frame = +2
Query: 326 NSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASNQIIGT 385
N+ GGIP+ L +L L L+SNK+S ++P L N S +L L LA N G
Sbjct: 5 NAFTGGIPEGMSRLHNLTFLSLASNKMSGEIPDDLFNCS------NLSTLSLAENNFSGL 166
Query: 386 V-PDMSGFSSLENMFLYENLLNGTI 409
+ PD+ L + L+ N G I
Sbjct: 167 IKPDIQNLLKLSRLQLHTNSFTGLI 241
>TC10104 similar to UP|Q8GTD6 (Q8GTD6) Polygalacturonase inhibitor-like
protein (Fragment), partial (42%)
Length = 563
Score = 44.3 bits (103), Expect = 5e-05
Identities = 31/92 (33%), Positives = 46/92 (49%), Gaps = 1/92 (1%)
Frame = +2
Query: 321 LIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASN 380
L I ++LEG IP +FG +LDLS N L +P + + + + L L+ N
Sbjct: 146 LNISHHALEGAIPDAFGERSYFTALDLSYNNLKGAIPKSISSAAY------IGHLDLSHN 307
Query: 381 QIIGTVPDMSGFSSLE-NMFLYENLLNGTILK 411
+ G +P S F LE + F+Y + L G LK
Sbjct: 308 HLCGAIPIGSPFDHLEASSFVYNDCLCGKPLK 403
Score = 32.7 bits (73), Expect = 0.14
Identities = 16/39 (41%), Positives = 23/39 (58%)
Frame = +2
Query: 320 SLIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPV 358
+L + N+L+G IPKS S + LDLS N L +P+
Sbjct: 215 ALDLSYNNLKGAIPKSISSAAYIGHLDLSHNHLCGAIPI 331
>TC9224 similar to GB|AAP21230.1|30102624|BT006422 At3g49750 {Arabidopsis
thaliana;}, partial (77%)
Length = 1318
Score = 44.3 bits (103), Expect = 5e-05
Identities = 38/134 (28%), Positives = 64/134 (47%), Gaps = 1/134 (0%)
Frame = +1
Query: 319 HSLIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLA 378
+ L + + +L G I + +L++LDLSSN L+ +P L +L +L L L+
Sbjct: 562 YKLSLNNLALHGTISPFLANCTNLQALDLSSNFLTGPIPPDLQSLV------NLAVLNLS 723
Query: 379 SNQIIGTVPDMSGFSSLENMF-LYENLLNGTILKNSTFPYRLANLYLDSNDLDGVITDSH 437
+N++ G +P + N+ L++NLL G I + RL+ + +N L G I S
Sbjct: 724 ANRLEGEIPPQLTMCAYLNIIDLHQNLLTGPIPQQLGLLVRLSAFDVSNNRLAGPIPSSL 903
Query: 438 FGNMSMLKYLSLSS 451
L + SS
Sbjct: 904 TNRSGNLPRFNASS 945
>TC16973 weakly similar to UP|Q96477 (Q96477) LRR protein, partial (30%)
Length = 555
Score = 43.9 bits (102), Expect = 6e-05
Identities = 29/80 (36%), Positives = 42/80 (52%)
Frame = +1
Query: 332 IPKSFGSLCSLRSLDLSSNKLSEDLPVMLHNLSVGCAKNSLKELYLASNQIIGTVPDMSG 391
IP SF +L SL+ L L++NKL+ +P L +L +LK +++N + GT+P
Sbjct: 1 IPNSFANLNSLKFLRLNNNKLTGSIPRELTHLK------NLKIFDVSNNDLCGTIPVDGN 162
Query: 392 FSSLENMFLYENLLNGTILK 411
F S N LNG LK
Sbjct: 163 FGSFPAESFENNQLNGPELK 222
Score = 29.6 bits (65), Expect = 1.2
Identities = 14/38 (36%), Positives = 22/38 (57%)
Frame = +1
Query: 321 LIIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLPV 358
L + +N L G IP+ L +L+ D+S+N L +PV
Sbjct: 40 LRLNNNKLTGSIPRELTHLKNLKIFDVSNNDLCGTIPV 153
>BP051648
Length = 558
Score = 42.4 bits (98), Expect = 2e-04
Identities = 40/145 (27%), Positives = 64/145 (43%), Gaps = 22/145 (15%)
Frame = +1
Query: 322 IIKSNSLEGGIPKSFGSLCSLRSLDLSSNKLSEDLP----------VMLHNLSVGCAKNS 371
I+++ S G + L S+R +D+S N ++ +P + N+S+ + S
Sbjct: 115 IVRNGSFTGTFKPPWHPLPSMRRIDVSDNVITGQIPSKNISSIFPNLRFLNMSINEIQGS 294
Query: 372 LKELY----------LASNQIIGTVP-DMSGF-SSLENMFLYENLLNGTILKNSTFPYRL 419
+ L+ N + G +P ++SG SSL + L N LNG + + L
Sbjct: 295 ISHELGQMKLLDTWDLSDNYLSGEIPKNISGDRSSLRFLRLSNNKLNGPVFPTWSALKHL 474
Query: 420 ANLYLDSNDLDGVITDSHFGNMSML 444
LYLD N G I S F N S+L
Sbjct: 475 EQLYLDGNSFSGSI-PSGFSNTSLL 546
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.338 0.146 0.471
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,585,398
Number of Sequences: 28460
Number of extensions: 147616
Number of successful extensions: 1332
Number of sequences better than 10.0: 85
Number of HSP's better than 10.0 without gapping: 1299
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1323
length of query: 491
length of database: 4,897,600
effective HSP length: 94
effective length of query: 397
effective length of database: 2,222,360
effective search space: 882276920
effective search space used: 882276920
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.8 bits)
S2: 57 (26.6 bits)
Medicago: description of AC136507.3


