

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136507.13 - phase: 0
(523 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
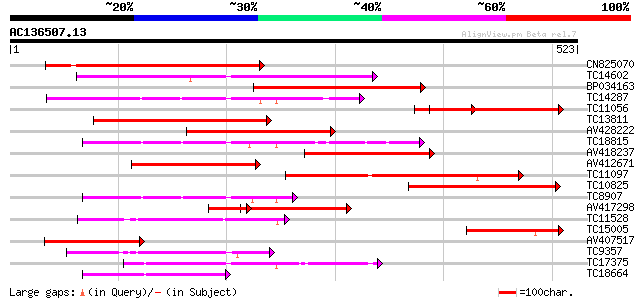
Score E
Sequences producing significant alignments: (bits) Value
CN825070 243 6e-65
TC14602 UP|Q8GS56 (Q8GS56) Phosphoenolpyruvate carboxylase kinas... 212 1e-55
BP034163 204 2e-53
TC14287 similar to UP|Q9LEU7 (Q9LEU7) Serine/threonine protein k... 182 1e-46
TC11056 similar to UP|O81390 (O81390) Calcium-dependent protein ... 180 6e-46
TC13811 homologue to UP|O24431 (O24431) Calmodulin-like domain p... 176 1e-44
AV428222 166 7e-42
TC18815 similar to UP|Q8W1D5 (Q8W1D5) CBL-interacting protein ki... 166 7e-42
AV418237 165 1e-41
AV412671 164 3e-41
TC11097 similar to UP|P93520 (P93520) Calcium/calmodulin-depende... 155 2e-38
TC10825 similar to GB|AAP68339.1|31711966|BT008900 At1g74740 {Ar... 150 4e-37
TC8907 similar to UP|Q8H0X3 (Q8H0X3) Serine/threonine protein ki... 141 2e-34
AV417298 128 2e-30
TC11528 homologue to UP|Q39868 (Q39868) Protein kinase , partial... 119 2e-27
TC15005 homologue to UP|Q9AR92 (Q9AR92) Protein kinase , partia... 108 2e-24
AV407517 104 3e-23
TC9357 homologue to UP|Q39193 (Q39193) Protein kinase (Protein k... 104 3e-23
TC17375 mitogen-activated kinase kinase kinase alpha [Lotus corn... 102 1e-22
TC18664 similar to UP|Q944I6 (Q944I6) At2g30360/T9D9.17, partial... 99 2e-21
>CN825070
Length = 634
Score = 243 bits (620), Expect = 6e-65
Identities = 113/202 (55%), Positives = 150/202 (73%)
Frame = +3
Query: 34 PPPWSKPSPQPSKPSKPSAIGPVLGRPMEDVKATYSMGKELGRGQFGVTHLCTHKTTGKQ 93
P P + P + P KP+ + P L E+++ Y+ G++LG+GQFG T+LC H +TG
Sbjct: 36 PVPATMSKPTTTAPPKPTTVLPYL---TENIREVYTFGRKLGQGQFGTTYLCRHNSTGCT 206
Query: 94 YACKTIAKRKLANKEDIEDVRREVQIMHHLTGQPNIVELIGAFEDKQSVHLVMELCAGGE 153
+ACK+I KRKL KED +DV RE+QIMHHL+ P++V + G +ED SVH+VME+C GGE
Sbjct: 207 FACKSIPKRKLLCKEDYDDVWREIQIMHHLSEHPHVVRIHGTYEDAASVHIVMEICEGGE 386
Query: 154 LFDRIIAKGHYTERAAASLLRTIVQIVHTCHSMGVIHRDLKPENFLLLSKDENSPLKATD 213
LFDRI+ KG Y+ER AA L+RTIV++V CHS+GV+HRDLKPENFL + +E++ LK TD
Sbjct: 387 LFDRIVQKGQYSEREAAKLIRTIVEVVEACHSLGVMHRDLKPENFLFDTVEEDAKLKTTD 566
Query: 214 FGLSVFYKQGDQFKDIVGSAYY 235
FGLSVFYK G+ F D+VGS YY
Sbjct: 567 FGLSVFYKPGETFGDVVGSPYY 632
>TC14602 UP|Q8GS56 (Q8GS56) Phosphoenolpyruvate carboxylase kinase, complete
Length = 1256
Score = 212 bits (540), Expect = 1e-55
Identities = 111/281 (39%), Positives = 169/281 (59%), Gaps = 3/281 (1%)
Frame = +2
Query: 62 EDVKATYSMGKELGRGQFGVTHLCTHKTTGKQYACKTIAKRKLANKEDIEDVRREVQIMH 121
E +K+ Y + +E+GRG+FG C H + +A K I K LA+ D + E + M
Sbjct: 50 EALKSQYQLCEEIGRGRFGTIFRCFHPISTDTFAVKLIDKSLLADSTDRHCLENEPKYMS 229
Query: 122 HLTGQPNIVELIGAFEDKQSVHLVMELCAGGELFDRIIAKGHYT--ERAAASLLRTIVQI 179
L+ PNI+++ FED + +V+ELC L DRI+A + E AA L++ +++
Sbjct: 230 LLSPHPNILQIFDVFEDDDVLSMVIELCQPLTLLDRIVAANGTSIPEVEAAGLMKQLLEA 409
Query: 180 VHTCHSMGVIHRDLKPENFLLLSKDENSPLKATDFGLSVFYKQGDQFKDIVGSAYYIAPD 239
V CH +GV HRD+KP+N L LK DFG + ++ G + +VG+ YY+AP+
Sbjct: 410 VAHCHRLGVAHRDVKPDNVLF---GGGGDLKLADFGSAEWFGDGRRMSGVVGTPYYVAPE 580
Query: 240 VLK-RKYGPEVDIWSVGVMLYILLCGVPPFWAESENGIFNAILRGHVDFSSDPWPSISPS 298
VL R+YG +VD+WS GV+LYI+L G PPF+ +S IF A++RG++ F S + ++SP+
Sbjct: 581 VLMGREYGEKVDVWSCGVILYIMLSGTPPFYGDSAAEIFEAVIRGNLRFPSRIFRNVSPA 760
Query: 299 AKDLVRKMLNSDPKQRITAYEVLNHPWIKEDGEAPDTPLDN 339
AKDL+RKM+ DP RI+A + L HPW G+ + +N
Sbjct: 761 AKDLLRKMICRDPSNRISAEQALRHPWFLSAGDTVNVT*EN 883
>BP034163
Length = 477
Score = 204 bits (520), Expect = 2e-53
Identities = 97/158 (61%), Positives = 120/158 (75%)
Frame = +1
Query: 226 FKDIVGSAYYIAPDVLKRKYGPEVDIWSVGVMLYILLCGVPPFWAESENGIFNAILRGHV 285
F +I GS YY+AP+VL+R YGPEVDIWS GV+LYILLCGVPPFWAE+E + AI+R V
Sbjct: 4 FNEIXGSPYYMAPEVLRRHYGPEVDIWSAGVILYILLCGVPPFWAETEQVVAQAIIRSVV 183
Query: 286 DFSSDPWPSISPSAKDLVRKMLNSDPKQRITAYEVLNHPWIKEDGEAPDTPLDNAVLNRL 345
DF DPWP +S +AKDLV+KMLN DPK+R+TA EVL+HPW+ +AP+ L V RL
Sbjct: 184 DFKRDPWPKVSDNAKDLVKKMLNPDPKRRLTAQEVLDHPWLVHAKKAPNVSLGETVKARL 363
Query: 346 KQFRAMNQFKKVALKVIASCLSEEEIMGLKQMFKGMDT 383
KQF MN+ KK AL+VIA L+ EE GL++ F+GMDT
Sbjct: 364 KQFSVMNKLKKRALRVIAEHLTVEEAAGLREGFQGMDT 477
>TC14287 similar to UP|Q9LEU7 (Q9LEU7) Serine/threonine protein kinase-like
protein (CBL-interacting protein kinase 5), partial
(75%)
Length = 1999
Score = 182 bits (462), Expect = 1e-46
Identities = 114/298 (38%), Positives = 158/298 (52%), Gaps = 5/298 (1%)
Frame = +3
Query: 35 PPWSKPSPQPSKPSKPSAIGPVLGRPMEDVKATYSMGKELGRGQFGVTHLCTHKTTGKQY 94
P + P P P P PS+ V P + Y MG+ LG+G F + + T +
Sbjct: 333 P*LATPPPPPPPPPPPSSSTMVPENPRNIIFNKYEMGRVLGQGNFAKVYHGRNLATNENV 512
Query: 95 ACKTIAKRKLANKEDIEDVRREVQIMHHLTGQPNIVELIGAFEDKQSVHLVMELCAGGEL 154
A K I K KL ++ ++REV +M L P+IVEL K + LVME GGEL
Sbjct: 513 AIKVIKKEKLKKDRLVKQIKREVSVM-RLVRHPHIVELKEVMATKGKIFLVMEYVKGGEL 689
Query: 155 FDRIIAKGHYTERAAASLLRTIVQIVHTCHSMGVIHRDLKPENFLLLSKDENSPLKATDF 214
F + + KG E A + ++ V CHS GV HRDLKPEN LL DEN LK +DF
Sbjct: 690 FTK-VNKGKLNEDDARKYFQQLISAVDFCHSRGVTHRDLKPENLLL---DENEDLKVSDF 857
Query: 215 GLSVFYKQGDQFKDIV---GSAYYIAPDVLKRK--YGPEVDIWSVGVMLYILLCGVPPFW 269
GLS +Q +V G+ Y+AP+VLK+K G + DIWS GV+LY LL G PF
Sbjct: 858 GLSALPEQRRDDGMLVTPCGTPAYVAPEVLKKKGYDGSKADIWSCGVILYALLSGYLPFQ 1037
Query: 270 AESENGIFNAILRGHVDFSSDPWPSISPSAKDLVRKMLNSDPKQRITAYEVLNHPWIK 327
E+ I+ + +F W ISP AK+L+ +L +DP++R + E+++ PW +
Sbjct: 1038GENVMRIYRKAFKAEYEFPE--W--ISPQAKNLISNLLVADPEKRYSIPEIISDPWFQ 1199
>TC11056 similar to UP|O81390 (O81390) Calcium-dependent protein kinase ,
partial (22%)
Length = 675
Score = 180 bits (456), Expect = 6e-46
Identities = 89/124 (71%), Positives = 103/124 (82%)
Frame = +3
Query: 388 TITIEELKQGLAKQGTRLSETEVKQLMEAADADGNGIIDYDEFITATMHMNRLNREEHVY 447
TIT EELK GLA+ G+RLSETEVKQLMEAAD DGNG IDY EFI+ATMH +RL R+EH+Y
Sbjct: 3 TITYEELKSGLARIGSRLSETEVKQLMEAADVDGNGSIDYLEFISATMHRHRLERDEHLY 182
Query: 448 TAFQFFDKDNSGYITIEELEQALHEYNMHDGRDIKEIISEVDADNDGRINYDEFVAMMGK 507
AFQ+FDKDNSG+ITI+ELE A+ ++ M D IKEIISEVD DNDGRINY+EF AMM
Sbjct: 183 KAFQYFDKDNSGHITIDELETAMTQHGMGDEASIKEIISEVDTDNDGRINYEEFCAMMRS 362
Query: 508 GNPE 511
G P+
Sbjct: 363 GMPQ 374
Score = 45.8 bits (107), Expect = 2e-05
Identities = 22/57 (38%), Positives = 35/57 (60%)
Frame = +3
Query: 374 LKQMFKGMDTDNSGTITIEELKQGLAKQGTRLSETEVKQLMEAADADGNGIIDYDEF 430
L + F+ D DNSG ITI+EL+ + + G E +K+++ D D +G I+Y+EF
Sbjct: 177 LYKAFQYFDKDNSGHITIDELETAMTQHGMG-DEASIKEIISEVDTDNDGRINYEEF 344
>TC13811 homologue to UP|O24431 (O24431) Calmodulin-like domain protein
kinase isoenzyme gamma , partial (27%)
Length = 573
Score = 176 bits (445), Expect = 1e-44
Identities = 91/164 (55%), Positives = 115/164 (69%)
Frame = +1
Query: 78 QFGVTHLCTHKTTGKQYACKTIAKRKLANKEDIEDVRREVQIMHHLTGQPNIVELIGAFE 137
QFGVT+LCT +T YACK+I+KRKL +K D ED++RE+QIM HL GQPNIVE GA+E
Sbjct: 1 QFGVTYLCTEISTKLLYACKSISKRKLVSKSDKEDIKREIQIMQHLIGQPNIVEFKGAYE 180
Query: 138 DKQSVHLVMELCAGGELFDRIIAKGHYTERAAASLLRTIVQIVHTCHSMGVIHRDLKPEN 197
DKQSVH+VMELCAGGELFDRIIAKGHY+ERAAAS+ R IV +VH CH MG +
Sbjct: 181 DKQSVHVVMELCAGGELFDRIIAKGHYSERAAASICRQIVNVVHVCHFMGCDA*GSEARE 360
Query: 198 FLLLSKDENSPLKATDFGLSVFYKQGDQFKDIVGSAYYIAPDVL 241
FL++ + S + F L F+ G F ++ + + I+P L
Sbjct: 361 FLII*QG*KSTSQGHGFWLVCFH**G*IFYPLIDNLFPISPAFL 492
>AV428222
Length = 416
Score = 166 bits (421), Expect = 7e-42
Identities = 76/137 (55%), Positives = 98/137 (71%)
Frame = +3
Query: 164 YTERAAASLLRTIVQIVHTCHSMGVIHRDLKPENFLLLSKDENSPLKATDFGLSVFYKQG 223
Y E A ++L I+ +V CH GV+HRDLKPENFL +SK+ +S +K DFGLS F +
Sbjct: 6 YPEDDAKAILLQILNVVAFCHLHGVVHRDLKPENFLFVSKEADSVMKVIDFGLSDFVRPD 185
Query: 224 DQFKDIVGSAYYIAPDVLKRKYGPEVDIWSVGVMLYILLCGVPPFWAESENGIFNAILRG 283
+ DIVGSAYY+AP+VL R Y E D+WS+GV+ YILLCG PFWA +E+GIF ++LR
Sbjct: 186 QRLNDIVGSAYYVAPEVLHRSYSVEADLWSIGVISYILLCGSRPFWARTESGIFRSVLRA 365
Query: 284 HVDFSSDPWPSISPSAK 300
+ +F PWPSISP AK
Sbjct: 366 NPNFDDSPWPSISPEAK 416
>TC18815 similar to UP|Q8W1D5 (Q8W1D5) CBL-interacting protein kinase
CIPK25, partial (58%)
Length = 979
Score = 166 bits (421), Expect = 7e-42
Identities = 108/320 (33%), Positives = 164/320 (50%), Gaps = 5/320 (1%)
Frame = +2
Query: 68 YSMGKELGRGQFGVTHLCTHKTTGKQYACKTIAKRKLANKEDIEDVRREVQIMHHLTGQP 127
Y MG+ LG+G + T+G+ A K ++K ++ + ++ ++RE+ IM L P
Sbjct: 38 YEMGRVLGKGTLAKVYFAKEITSGEGVAIKVMSKARIKKEGMMDQIKREISIMR-LVRHP 214
Query: 128 NIVELIGAFEDKQSVHLVMELCAGGELFDRIIAKGHYTERAAASLLRTIVQIVHTCHSMG 187
NIV L K + +ME GGELF ++ AKG + A + ++ V CHS G
Sbjct: 215 NIVNLKEVMATKTKIFFIMEYIRGGELFAKV-AKGKLKDDLARRYFQQLISAVDYCHSRG 391
Query: 188 VIHRDLKPENFLLLSKDENSPLKATDFGLSVF---YKQGDQFKDIVGSAYYIAPDVLKRK 244
V HRDLKPEN LL DEN LK +DFGLS +Q G+ Y+AP+VL++K
Sbjct: 392 VSHRDLKPENLLL---DENENLKVSDFGLSGLPEQLRQDGLLHTQCGTPAYVAPEVLRKK 562
Query: 245 --YGPEVDIWSVGVMLYILLCGVPPFWAESENGIFNAILRGHVDFSSDPWPSISPSAKDL 302
G + D WS GV+LY LL G PF E+ ++N +LR +F PW SP +K L
Sbjct: 563 GYDGFKTDTWSCGVILYALLAGCLPFQHENLMTMYNKVLR--AEFQFPPW--FSPESKKL 730
Query: 303 VRKMLNSDPKQRITAYEVLNHPWIKEDGEAPDTPLDNAVLNRLKQFRAMNQFKKVALKVI 362
+ K+L +DP +RIT ++ W ++ G + P+ + + +N + + KV+
Sbjct: 731 ISKILVADPNRRITISSIMRVSWFQK-GFSASIPIPDPDESNFNS--DLNSSSEQSTKVV 901
Query: 363 ASCLSEEEIMGLKQMFKGMD 382
A+ + M G D
Sbjct: 902 AAAKFINAFEFISSMSSGFD 961
>AV418237
Length = 361
Score = 165 bits (418), Expect = 1e-41
Identities = 80/120 (66%), Positives = 98/120 (81%)
Frame = +2
Query: 273 ENGIFNAILRGHVDFSSDPWPSISPSAKDLVRKMLNSDPKQRITAYEVLNHPWIKEDGEA 332
E GIF IL G +DF S+PWPSIS SAKDL+RKML+ +PK R+TA++VL HPWI +D A
Sbjct: 2 EQGIFRQILMGRLDFQSEPWPSISDSAKDLIRKMLDRNPKTRLTAHQVLCHPWIVDDNIA 181
Query: 333 PDTPLDNAVLNRLKQFRAMNQFKKVALKVIASCLSEEEIMGLKQMFKGMDTDNSGTITIE 392
PD PLD+AVL+RLKQF AMN+ KK+AL+VIA LSEEEI GLK++FK +D DNSGTIT +
Sbjct: 182 PDKPLDSAVLSRLKQFSAMNKLKKMALRVIAESLSEEEIGGLKELFKMIDADNSGTITFD 361
>AV412671
Length = 359
Score = 164 bits (415), Expect = 3e-41
Identities = 78/119 (65%), Positives = 94/119 (78%)
Frame = +3
Query: 113 VRREVQIMHHLTGQPNIVELIGAFEDKQSVHLVMELCAGGELFDRIIAKGHYTERAAASL 172
VRREV+IM HL PNIV L +ED +VHLVMELC GGELFDRI+A+GHYTERAAA++
Sbjct: 3 VRREVEIMRHLPRHPNIVWLKDTYEDDNAVHLVMELCEGGELFDRIVARGHYTERAAAAV 182
Query: 173 LRTIVQIVHTCHSMGVIHRDLKPENFLLLSKDENSPLKATDFGLSVFYKQGDQFKDIVG 231
+TIV++V CH GV+HRDLKPENFL +K E + LKA DFGLSVF+K G+ F +IVG
Sbjct: 183 TKTIVEVVQMCHKHGVMHRDLKPENFLFANKKETAALKAIDFGLSVFFKPGETFNEIVG 359
>TC11097 similar to UP|P93520 (P93520) Calcium/calmodulin-dependent protein
kinase homolog|CaM kinase homolog|MCK1, partial (36%)
Length = 949
Score = 155 bits (391), Expect = 2e-38
Identities = 78/225 (34%), Positives = 137/225 (60%), Gaps = 5/225 (2%)
Frame = +2
Query: 255 GVMLYILLCGVPPFWAESENGIFNAILRGHVDFSSDPWPSISPSAKDLVRKMLNSDPKQR 314
GV+ YILLCG PFWA +E+GIF A+LR +F PWPS++P AKD V+++LN D ++R
Sbjct: 20 GVITYILLCGSRPFWARTESGIFRAVLRADPNFDDLPWPSVTPEAKDFVKRLLNKDYRKR 199
Query: 315 ITAYEVLNHPWIKEDGEAPDTPLDNAVLNRLKQFRAMNQFKKVALKVIASCLSEEEIMGL 374
+TA + L HPW+++D PLD + +K + FK+ A+K ++ L+EE+++ L
Sbjct: 200 MTAAQALTHPWLRDDSR--PIPLDILIYKLVKSYLHATPFKRAAVKALSKALTEEQLVYL 373
Query: 375 KQMFKGMDTDNSGTITIEELKQGLAKQGT-RLSETEVKQLMEAADADGNGIIDYDEF--- 430
+ F+ ++ + G I+++ K LA+ T + E+ V ++ + +D++EF
Sbjct: 374 RAQFRLLEPNRDGHISLDNFKMALARHATDAMRESRVLDIIHMMEPLAYRKMDFEEFCAA 553
Query: 431 ITATMHMNRLNREEHVYT-AFQFFDKDNSGYITIEELEQALHEYN 474
T+T + L+R E + + AF+ F+++ + I+IEEL + L ++
Sbjct: 554 ATSTYQLEALDRWEDIASAAFEHFEREGNRVISIEELARELKSWS 688
>TC10825 similar to GB|AAP68339.1|31711966|BT008900 At1g74740 {Arabidopsis
thaliana;}, partial (32%)
Length = 1091
Score = 150 bits (380), Expect = 4e-37
Identities = 72/141 (51%), Positives = 99/141 (70%), Gaps = 1/141 (0%)
Frame = +1
Query: 369 EEIMGLKQMFKGMDTDNSGTITIEELKQGLAKQGTRLSETEVKQLMEAADADGNGIIDYD 428
EE+ +K MF MDTD G ++ EELK GL K G++L++ E+K LME AD DGNG++DY
Sbjct: 4 EEVEIIKDMFTLMDTDKDGRVSYEELKAGLRKVGSQLADQEIKMLMEVADVDGNGVLDYG 183
Query: 429 EFITATMHMNRLNREEHVYTAFQFFDKDNSGYITIEELEQAL-HEYNMHDGRDIKEIISE 487
EF+ T+H+ ++ +EH + AF+FFDKD SGYI EL++AL E + D + +I+ E
Sbjct: 184 EFVAVTIHLQKMENDEHFHKAFKFFDKDGSGYIESGELQEALADESGVTDADVLNDIMRE 363
Query: 488 VDADNDGRINYDEFVAMMGKG 508
VD D DGRI+Y+EFVAMM G
Sbjct: 364 VDTDKDGRISYEEFVAMMKAG 426
>TC8907 similar to UP|Q8H0X3 (Q8H0X3) Serine/threonine protein kinase-like
protein, partial (44%)
Length = 941
Score = 141 bits (356), Expect = 2e-34
Identities = 84/203 (41%), Positives = 115/203 (56%), Gaps = 5/203 (2%)
Frame = +3
Query: 68 YSMGKELGRGQFGVTHLCTHKTTGKQYACKTIAKRKLANKEDIEDVRREVQIMHHLTGQP 127
Y +GK LG+G F + + T + A K I K +L + ++ ++REV +M L P
Sbjct: 342 YEIGKILGQGNFAKVYHGRNMETNESVAIKVIKKERLKKERLVKQIKREVSVMR-LVRHP 518
Query: 128 NIVELIGAFEDKQSVHLVMELCAGGELFDRIIAKGHYTERAAASLLRTIVQIVHTCHSMG 187
+IVEL K + +V+E GGELF ++ KG TE AA + ++ V CHS G
Sbjct: 519 HIVELKEVMATKTKIFMVVEYVKGGELFAKL-TKGKMTEVAARKYFQQLISAVDFCHSRG 695
Query: 188 VIHRDLKPENFLLLSKDENSPLKATDFGLSVFYKQ---GDQFKDIVGSAYYIAPDVLKRK 244
V HRDLKPEN LL D+N LK +DFGLS +Q G+ Y+AP+VLK+K
Sbjct: 696 VTHRDLKPENLLL---DDNEDLKVSDFGLSSLPEQRRSDGMLLTPCGTPAYVAPEVLKKK 866
Query: 245 --YGPEVDIWSVGVMLYILLCGV 265
G + DIWS GV+LY LLCG+
Sbjct: 867 GYDGSKADIWSCGVILYALLCGI 935
>AV417298
Length = 424
Score = 128 bits (322), Expect = 2e-30
Identities = 58/102 (56%), Positives = 77/102 (74%)
Frame = +1
Query: 214 FGLSVFYKQGDQFKDIVGSAYYIAPDVLKRKYGPEVDIWSVGVMLYILLCGVPPFWAESE 273
+ LS+ G +F+DIVGSAYY+AP+VLKRK GPE D+WS+GV+ YILLCG PFW ++E
Sbjct: 118 WSLSLKTISGKKFQDIVGSAYYVAPEVLKRKSGPESDVWSIGVITYILLCGRRPFWDKTE 297
Query: 274 NGIFNAILRGHVDFSSDPWPSISPSAKDLVRKMLNSDPKQRI 315
+GIF +LR DF PWP+IS +AKD V+K+L DP+ R+
Sbjct: 298 DGIFKEVLRNKPDFRRKPWPTISNAAKDFVKKLLVKDPRARL 423
Score = 55.8 bits (133), Expect = 2e-08
Identities = 26/40 (65%), Positives = 30/40 (75%)
Frame = +2
Query: 184 HSMGVIHRDLKPENFLLLSKDENSPLKATDFGLSVFYKQG 223
H G++HRD+KPENFL S E+S LKATDFGLS F K G
Sbjct: 2 HLHGLVHRDMKPENFLFKSNREDSALKATDFGLSDFIKPG 121
>TC11528 homologue to UP|Q39868 (Q39868) Protein kinase , partial (56%)
Length = 640
Score = 119 bits (297), Expect = 2e-27
Identities = 78/198 (39%), Positives = 104/198 (52%), Gaps = 2/198 (1%)
Frame = +3
Query: 63 DVKATYSMGKELGRGQFGVTHLCTHKTTGKQYACKTIAKRKLANKEDIEDVRREVQIMHH 122
D++ Y KELG G FGV L K TG+ A K I + K + E+V+RE+ I H
Sbjct: 63 DMEERYEPLKELGSGNFGVARLARDKNTGELVAVKYIERGKKID----ENVQREI-INHR 227
Query: 123 LTGQPNIVELIGAFEDKQSVHLVMELCAGGELFDRIIAKGHYTERAAASLLRTIVQIVHT 182
PNI+ + +V+E +GGELF+RI + G ++E A + ++ V
Sbjct: 228 SLRHPNIIRFKEVLLTPTHLAIVLEYASGGELFERICSAGRFSEDEARYFFQQLISGVSY 407
Query: 183 CHSMGVIHRDLKPENFLLLSKDENSPLKATDFGLSVFYKQGDQFKDIVGSAYYIAPDVLK 242
CHSM + HRDLK EN LL + + LK DFG S Q K VG+ YIAP+VL
Sbjct: 408 CHSMEICHRDLKLEN-TLLDGNPSPRLKICDFGYSKSAILHSQPKSTVGTPAYIAPEVLS 584
Query: 243 RKY--GPEVDIWSVGVML 258
RK G D+WS GV L
Sbjct: 585 RKEYDGKVADVWSCGVTL 638
>TC15005 homologue to UP|Q9AR92 (Q9AR92) Protein kinase , partial (19%)
Length = 657
Score = 108 bits (271), Expect = 2e-24
Identities = 55/97 (56%), Positives = 70/97 (71%), Gaps = 7/97 (7%)
Frame = +2
Query: 422 NGIIDYDEFITATMHMNRLNREEHVYTAFQFFDKDNSGYITIEELEQALHEYNMHDGRDI 481
+G IDY EF+TATMH ++L R++H+ AF +FDKD+SG+IT +ELE A+ EY M D I
Sbjct: 14 DGTIDYIEFVTATMHRHKLERDDHLN*AFPYFDKDSSGFITRDELEIAMKEYGMGDDATI 193
Query: 482 KE-------IISEVDADNDGRINYDEFVAMMGKGNPE 511
KE IISEVD D+DGRINY+EF AMM GN +
Sbjct: 194 KEIISEVDTIISEVDTDHDGRINYEEFCAMMRSGNQQ 304
>AV407517
Length = 424
Score = 104 bits (260), Expect = 3e-23
Identities = 51/93 (54%), Positives = 66/93 (70%), Gaps = 1/93 (1%)
Frame = +2
Query: 33 TPPPWSKPS-PQPSKPSKPSAIGPVLGRPMEDVKATYSMGKELGRGQFGVTHLCTHKTTG 91
TPP KPS PQ + + +LG+P ED+K Y++GKELGRGQFGVT+LCT +TG
Sbjct: 146 TPPQNPKPSTPQNAVRTVQKTEPTILGKPFEDIKNHYTLGKELGRGQFGVTYLCTENSTG 325
Query: 92 KQYACKTIAKRKLANKEDIEDVRREVQIMHHLT 124
YACK+I KRKL +K D ED++RE+ IM HL+
Sbjct: 326 NFYACKSILKRKLVSKADKEDMKREIHIMQHLS 424
>TC9357 homologue to UP|Q39193 (Q39193) Protein kinase (Protein kinase,
41K) , partial (71%)
Length = 1181
Score = 104 bits (260), Expect = 3e-23
Identities = 73/195 (37%), Positives = 100/195 (50%), Gaps = 3/195 (1%)
Frame = +1
Query: 53 IGPVLGRPMEDVKATYSMGKELGRGQFGVTHLCTHKTTGKQYACKTIAKRKLANKEDIED 112
+GP + P+ Y + +++G G FGV L K T + A K I + +K D E+
Sbjct: 316 VGPGMDMPIMHDSDRYDLVRDIGSGNFGVARLMQDKQTKELVAVKYIER---GDKID-EN 483
Query: 113 VRREVQIMHHLTGQPNIVELIGAFEDKQSVHLVMELCAGGELFDRIIAKGHYTERAAASL 172
V+RE+ I H PNIV + +VME +GGELF+RI G +TE A
Sbjct: 484 VKREI-INHRSLRHPNIVRFKEVILTPTHLAIVMEYASGGELFERICNAGRFTEDEARFF 660
Query: 173 LRTIVQIVHTCHSMGVIHRDLKPENFLLLSKDENSP---LKATDFGLSVFYKQGDQFKDI 229
+ ++ V CH+M V HRDLK EN LL + SP LK DFG S Q K
Sbjct: 661 FQQLISGVSYCHAMQVCHRDLKLENTLL----DGSPTPRLKICDFGYSKSSVLHSQPKST 828
Query: 230 VGSAYYIAPDVLKRK 244
VG+ YIAP+VL ++
Sbjct: 829 VGTPAYIAPEVLSKQ 873
>TC17375 mitogen-activated kinase kinase kinase alpha [Lotus corniculatus
var. japonicus]
Length = 1422
Score = 102 bits (255), Expect = 1e-22
Identities = 69/241 (28%), Positives = 120/241 (49%), Gaps = 2/241 (0%)
Frame = +3
Query: 106 NKEDIEDVRREVQIMHHLTGQPNIVELIGAFEDKQSVHLVMELCAGGELFDRIIAKGHYT 165
+KE ++ + +E+ +++ + PNIV+ G+ ++S+ + +E +GG + + G +
Sbjct: 51 SKECLKQLNQEINLLNQFS-HPNIVQYYGSELGEESLSVYLEYVSGGSIHKLLQEYGAFK 227
Query: 166 ERAAASLLRTIVQIVHTCHSMGVIHRDLKPENFLLLSKDENSPLKATDFGLSVFYKQGDQ 225
E + R IV + HS +HRD+K N L+ D N +K DFG+S
Sbjct: 228 EPVIQNYTRQIVSGLAYLHSRNTVHRDIKGANILV---DPNGEIKLADFGMSKHINSAAS 398
Query: 226 FKDIVGSAYYIAPDVLKRK--YGPEVDIWSVGVMLYILLCGVPPFWAESENGIFNAILRG 283
GS Y++AP+V+ YG VDI S+G + + PP W++ E G+ G
Sbjct: 399 MLSFKGSPYWMAPEVVMNTNGYGLPVDISSLGCTILEMATSKPP-WSQFE-GVAAIFKIG 572
Query: 284 HVDFSSDPWPSISPSAKDLVRKMLNSDPKQRITAYEVLNHPWIKEDGEAPDTPLDNAVLN 343
+ + +S AK+ +++ L DP R TA +LNHP+I++ T + NA +
Sbjct: 573 NSKDMPEIPEHLSDDAKNFIKQCLQRDPLARPTAQSLLNHPFIRDQSA---TKVANASIT 743
Query: 344 R 344
R
Sbjct: 744 R 746
>TC18664 similar to UP|Q944I6 (Q944I6) At2g30360/T9D9.17, partial (34%)
Length = 643
Score = 98.6 bits (244), Expect = 2e-21
Identities = 57/136 (41%), Positives = 72/136 (52%)
Frame = +3
Query: 68 YSMGKELGRGQFGVTHLCTHKTTGKQYACKTIAKRKLANKEDIEDVRREVQIMHHLTGQP 127
Y +G+ LG G F + + TG+ A K I K+K+ V+REV IM L P
Sbjct: 93 YEVGRLLGCGAFAKVYHARNIETGQSVAVKVINKKKVIGTGLTGHVKREVSIMSRLR-HP 269
Query: 128 NIVELIGAFEDKQSVHLVMELCAGGELFDRIIAKGHYTERAAASLLRTIVQIVHTCHSMG 187
NIV L K ++ VME GGELF RI KG ++E A + ++ V CHS G
Sbjct: 270 NIVRLHEVLATKTKIYFVMEFAKGGELFARISTKGRFSEDLARRYFQQLISAVGYCHSRG 449
Query: 188 VIHRDLKPENFLLLSK 203
V HRDLKPEN LL K
Sbjct: 450 VFHRDLKPENLLLDDK 497
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.134 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,169,094
Number of Sequences: 28460
Number of extensions: 113365
Number of successful extensions: 1992
Number of sequences better than 10.0: 265
Number of HSP's better than 10.0 without gapping: 1578
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1858
length of query: 523
length of database: 4,897,600
effective HSP length: 94
effective length of query: 429
effective length of database: 2,222,360
effective search space: 953392440
effective search space used: 953392440
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 57 (26.6 bits)
Medicago: description of AC136507.13


