

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136506.11 - phase: 0 /pseudo
(1043 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
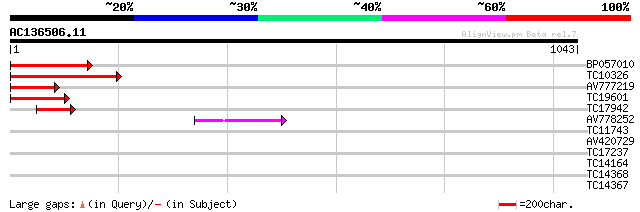
Score E
Sequences producing significant alignments: (bits) Value
BP057010 224 5e-59
TC10326 similar to AAQ89612 (AAQ89612) At5g26850, partial (21%) 222 2e-58
AV777219 179 3e-45
TC19601 similar to UP|Q94KD1 (Q94KD1) At1g05960/T21E18_20, parti... 167 1e-41
TC17942 weakly similar to UP|Q94KD1 (Q94KD1) At1g05960/T21E18_20... 86 4e-17
AV778252 50 2e-06
TC11743 similar to UP|Q94KD1 (Q94KD1) At1g05960/T21E18_20, parti... 34 0.10
AV420729 30 2.0
TC17237 weakly similar to UP|Q9SS82 (Q9SS82) MZB10.8 protein, pa... 29 4.4
TC14164 weakly similar to UP|Q8VYH6 (Q8VYH6) AT4g17280/dl4675c, ... 28 7.4
TC14368 similar to UP|Q9LLC2 (Q9LLC2) Xyloglucan endotransglycos... 28 9.7
TC14367 similar to UP|Q9ZRV1 (Q9ZRV1) Xyloglucan endotransglycos... 28 9.7
>BP057010
Length = 573
Score = 224 bits (571), Expect = 5e-59
Identities = 102/152 (67%), Positives = 124/152 (81%)
Frame = +2
Query: 1 MSVISRNIWPVCGSLCCFCPALRERSRHPIKRYKKLLADIFPRTPEEEPNDRKISKLCEY 60
M V+SR + PVCG+LCC CP+LR SR P+KRYKKLLADIFPR E EPNDRK+ KLC+Y
Sbjct: 116 MGVMSRRVVPVCGNLCCMCPSLRASSRQPVKRYKKLLADIFPRNQEAEPNDRKMGKLCDY 295
Query: 61 ASKNPLRVPKITSYLEQRCYKELRTENYQAVKVVICIYRKLLVSCRDQMPLFASSLLSII 120
ASKNPLR+PK+T LEQ CYK+LR E + +VKVV+CIYRK L SC++ MPLFA SLL II
Sbjct: 296 ASKNPLRIPKVTDNLEQICYKDLRNEVFGSVKVVLCIYRKFLSSCKEHMPLFAGSLLEII 475
Query: 121 QILLDQSRQDEVQILGCQTLFDFVNNQRDGTY 152
+ LL+Q+R DE++ILGC TL DF++ Q DGTY
Sbjct: 476 RTLLEQTRTDEIRILGCNTLIDFIDRQTDGTY 571
>TC10326 similar to AAQ89612 (AAQ89612) At5g26850, partial (21%)
Length = 746
Score = 222 bits (566), Expect = 2e-58
Identities = 105/205 (51%), Positives = 147/205 (71%)
Frame = +2
Query: 1 MSVISRNIWPVCGSLCCFCPALRERSRHPIKRYKKLLADIFPRTPEEEPNDRKISKLCEY 60
M +ISR I+P CG++C CPALR SR P+KRYKKLLADIFP++P+E P++RKI KLCEY
Sbjct: 131 MGIISRKIFPACGNMCVCCPALRSSSRQPVKRYKKLLADIFPKSPDELPSERKIVKLCEY 310
Query: 61 ASKNPLRVPKITSYLEQRCYKELRTENYQAVKVVICIYRKLLVSCRDQMPLFASSLLSII 120
A+KNP R+PKI YLE+RCYKELR+ + + V ++ + KLL C+ Q+ FA +L++I
Sbjct: 311 AAKNPFRIPKIAKYLEERCYKELRSGHIKLVTIITESFNKLLSICKVQIAYFALDVLNVI 490
Query: 121 QILLDQSRQDEVQILGCQTLFDFVNNQRDGTYMFNLDSFILKLCHLAQQVGDDGKVEHLR 180
LL S+ + +Q LGCQ+L F+ Q D TY N++ + K+C L+++ G+ + LR
Sbjct: 491 TELLGYSKDETIQTLGCQSLTRFIFCQVDTTYTHNIEKLVRKVCILSREHGETHEKRCLR 670
Query: 181 ASGLQVLSSMVWFMGEFTHISVEFD 205
AS LQ LS+MVWFM EF++I +FD
Sbjct: 671 ASSLQCLSAMVWFMAEFSNIFADFD 745
>AV777219
Length = 291
Score = 179 bits (453), Expect = 3e-45
Identities = 84/91 (92%), Positives = 87/91 (95%)
Frame = -2
Query: 1 MSVISRNIWPVCGSLCCFCPALRERSRHPIKRYKKLLADIFPRTPEEEPNDRKISKLCEY 60
MSVISRNIWPVCGSLC FCPALRERSRHPIKRYK LLADIFPRT EEEPNDRKISKLCEY
Sbjct: 275 MSVISRNIWPVCGSLCFFCPALRERSRHPIKRYKHLLADIFPRTQEEEPNDRKISKLCEY 96
Query: 61 ASKNPLRVPKITSYLEQRCYKELRTENYQAV 91
ASK+PLRVPKITSYLEQRCY+ELR ENYQ+V
Sbjct: 95 ASKDPLRVPKITSYLEQRCYRELRNENYQSV 3
>TC19601 similar to UP|Q94KD1 (Q94KD1) At1g05960/T21E18_20, partial (15%)
Length = 547
Score = 167 bits (422), Expect = 1e-41
Identities = 75/109 (68%), Positives = 89/109 (80%)
Frame = +2
Query: 1 MSVISRNIWPVCGSLCCFCPALRERSRHPIKRYKKLLADIFPRTPEEEPNDRKISKLCEY 60
M V+SR + PVCG+LCC CP+LR SR P+KRYKKLLADIFPR E EPNDRKI KLC+Y
Sbjct: 221 MGVMSRRVVPVCGNLCCMCPSLRASSRQPVKRYKKLLADIFPRNQEAEPNDRKIGKLCDY 400
Query: 61 ASKNPLRVPKITSYLEQRCYKELRTENYQAVKVVICIYRKLLVSCRDQM 109
ASKNPLR+PK+T LEQ CYK+LR E + +VKVV+CIYRK L SC++ M
Sbjct: 401 ASKNPLRIPKVTDNLEQICYKDLRNEVFGSVKVVLCIYRKFLSSCKEHM 547
>TC17942 weakly similar to UP|Q94KD1 (Q94KD1) At1g05960/T21E18_20, partial
(8%)
Length = 523
Score = 85.5 bits (210), Expect = 4e-17
Identities = 39/71 (54%), Positives = 53/71 (73%)
Frame = +2
Query: 50 NDRKISKLCEYASKNPLRVPKITSYLEQRCYKELRTENYQAVKVVICIYRKLLVSCRDQM 109
NDRKI KLC+YAS + L +P++ + LEQRCYKELR EN + K+V+CIY+K L SC++QM
Sbjct: 2 NDRKIGKLCDYASPHLLPIPQLVNALEQRCYKELRNENLHSTKIVMCIYKKFLFSCKEQM 181
Query: 110 PLFASSLLSII 120
++ LS I
Sbjct: 182 FSLWANTLSYI 214
>AV778252
Length = 522
Score = 50.1 bits (118), Expect = 2e-06
Identities = 48/172 (27%), Positives = 83/172 (47%), Gaps = 3/172 (1%)
Frame = -1
Query: 340 ECLKES*YAT*YCRCYYPSC*KNKSSAIGGYNWCIE*YDETPAEKHTLLSR*FELRNRSN 399
EC + + + *Y + Y+ +C K++++ NWC * T E +L F ++
Sbjct: 513 ECCQAAYSSN*YYQHYHTTCTKSEATDFSCNNWCYI*SH*TLTEVPAILI*SF--KHWKQ 340
Query: 400 TMEPKI*NGS--R*MPCTVNN*DLRCRSCSRYHGRVVRKHV*YYSYGKNLDCCSL*NFSN 457
+ K * R + T RCR+ S + G V +++ YY Y KN + + N
Sbjct: 339 WL*VKC*TSICLRNVYITTFKQAWRCRTHS*FDGCSVGEYLEYYHYSKNNNLRYISNCKI 160
Query: 458 SCIYTEFVVSEQGIP*GIISSVTPGNGPCRP*N-QGGCTPHIFSCSCAIVSL 508
+ I++ +S++G P* ++SS+ GNG P*+ + G + C C + SL
Sbjct: 159 NHIHS*CFISKEGFP*CLVSSIAHGNGSS*P*HPELGHIVYSLLCLCHLCSL 4
>TC11743 similar to UP|Q94KD1 (Q94KD1) At1g05960/T21E18_20, partial (3%)
Length = 416
Score = 34.3 bits (77), Expect = 0.10
Identities = 13/18 (72%), Positives = 14/18 (77%)
Frame = +1
Query: 3 VISRNIWPVCGSLCCFCP 20
VISR + P CGSLC FCP
Sbjct: 361 VISRQVLPACGSLCFFCP 414
>AV420729
Length = 208
Score = 30.0 bits (66), Expect = 2.0
Identities = 16/56 (28%), Positives = 23/56 (40%), Gaps = 9/56 (16%)
Frame = -2
Query: 12 CGSLCCFCPALRERSRHPIKRYKKLLADIFP---------RTPEEEPNDRKISKLC 58
C CC PA R HP+ ++ L +P + EE +D + SK C
Sbjct: 183 CKGFCCISPAARICKSHPVVQFSHSLPPSYP*YS*KWNWNKKGSEEISDEQESKRC 16
>TC17237 weakly similar to UP|Q9SS82 (Q9SS82) MZB10.8 protein, partial (18%)
Length = 712
Score = 28.9 bits (63), Expect = 4.4
Identities = 13/36 (36%), Positives = 23/36 (63%)
Frame = -3
Query: 89 QAVKVVICIYRKLLVSCRDQMPLFASSLLSIIQILL 124
Q+ I IY+ LL+ +D++ LF SSL+ + +L+
Sbjct: 377 QSSNSAIPIYKCLLIKIKDELELFLSSLIFFLLVLV 270
>TC14164 weakly similar to UP|Q8VYH6 (Q8VYH6) AT4g17280/dl4675c, partial
(44%)
Length = 1606
Score = 28.1 bits (61), Expect = 7.4
Identities = 11/25 (44%), Positives = 16/25 (64%)
Frame = -1
Query: 161 LKLCHLAQQVGDDGKVEHLRASGLQ 185
+ LCH+ VGD G V+ R +GL+
Sbjct: 490 IPLCHIGVVVGDGGSVDFRRTTGLR 416
>TC14368 similar to UP|Q9LLC2 (Q9LLC2) Xyloglucan endotransglycosylase XET2
, partial (46%)
Length = 565
Score = 27.7 bits (60), Expect = 9.7
Identities = 12/32 (37%), Positives = 17/32 (52%)
Frame = +3
Query: 208 SLASLVNSGCFGCFGKLWGYQGRFSKREFYRA 239
SL LV GC GC W ++R+F++A
Sbjct: 285 SLIPLVTRGCGGCRRTTWSTITALTRRDFHKA 380
>TC14367 similar to UP|Q9ZRV1 (Q9ZRV1) Xyloglucan endotransglycosylase 1,
partial (96%)
Length = 1137
Score = 27.7 bits (60), Expect = 9.7
Identities = 12/32 (37%), Positives = 17/32 (52%)
Frame = +1
Query: 208 SLASLVNSGCFGCFGKLWGYQGRFSKREFYRA 239
SL LV GC GC W ++R+F++A
Sbjct: 850 SLIPLVTRGCGGCRRTTWSTITALTRRDFHKA 945
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.361 0.161 0.605
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,362,360
Number of Sequences: 28460
Number of extensions: 314229
Number of successful extensions: 3394
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 2480
Number of HSP's successfully gapped in prelim test: 57
Number of HSP's that attempted gapping in prelim test: 856
Number of HSP's gapped (non-prelim): 2632
length of query: 1043
length of database: 4,897,600
effective HSP length: 100
effective length of query: 943
effective length of database: 2,051,600
effective search space: 1934658800
effective search space used: 1934658800
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 14 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC136506.11


