

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136504.2 + phase: 0
(713 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
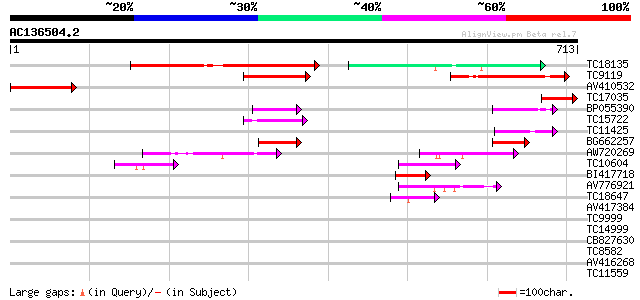
Score E
Sequences producing significant alignments: (bits) Value
TC18135 homologue to UP|Q9MAY4 (Q9MAY4) ABC transporter homolog,... 220 7e-58
TC9119 homologue to AAQ65167 (AAQ65167) At1g64550, partial (20%) 130 9e-31
AV410532 117 6e-27
TC17035 similar to UP|Q9M1H3 (Q9M1H3) ABC transporter-like prote... 95 4e-20
BP055390 47 8e-06
TC15722 similar to UP|Q9FMU0 (Q9FMU0) Similarity to ABC transpor... 47 1e-05
TC11425 similar to UP|Q9M1Q9 (Q9M1Q9) P-glycoprotein-like proeti... 45 3e-05
BG662257 45 4e-05
AW720269 44 7e-05
TC10604 similar to AAQ89671 (AAQ89671) At4g33460, partial (36%) 44 9e-05
BI417718 42 3e-04
AV776921 42 3e-04
TC18647 similar to UP|Q9ASR9 (Q9ASR9) At2g01320/F10A8.20, partia... 42 3e-04
AV417384 39 0.003
TC9999 similar to UP|Q9FMU0 (Q9FMU0) Similarity to ABC transport... 38 0.005
TC14999 similar to UP|Q9FJH6 (Q9FJH6) ABC transporter homolog Pn... 38 0.006
CB827630 37 0.008
TC8582 similar to UP|Q8LJS2 (Q8LJS2) Nucleolar histone deacetyla... 37 0.011
AV416268 37 0.011
TC11559 similar to UP|Q9SXT3 (Q9SXT3) Multidrug resistance prote... 37 0.011
>TC18135 homologue to UP|Q9MAY4 (Q9MAY4) ABC transporter homolog, partial
(50%)
Length = 968
Score = 220 bits (560), Expect = 7e-58
Identities = 107/238 (44%), Positives = 165/238 (68%)
Frame = +3
Query: 152 KDITIENFSVAARGKELLKNTSVKISHGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNID 211
+DI IE+ SV G +L+ ++ +++++G+RYGL+G NG GKSTLL + R++P+P+++D
Sbjct: 294 RDIRIESLSVTFHGHDLIVDSELELNYGRRYGLLGLNGCGKSTLLTAIGHRELPIPEHMD 473
Query: 212 VLLVEQEVVGDDKTALEAVVSANVELIKVRQKVADLQNIASGEEGMDKDDTNEEEDAGEK 271
+ + +E+ D +ALEAVVS + E +++ ++ + +A+ ++G GE
Sbjct: 474 IYHLTREIEASDMSALEAVVSCDEERLRLEKEA---ETLAAQDDG-----------GGET 611
Query: 272 LAELYEQLQLMGSDAAESQASKILAGLGFTKDMQGRPTKSFSGGWRMRISLARALFVQPT 331
L +YE+L+ + + AE +A++IL GLGF K MQ + T+ FSGGWRMRI+LARALF+ PT
Sbjct: 612 LERIYERLEALDASTAEKRAAEILHGLGFDKKMQAKKTRDFSGGWRMRIALARALFINPT 791
Query: 332 LLLLDEPTNHLDLRAVLWLEEYLCRWKKTLVVVSHDRDFLNTVCSEIIHLHDLKLHFY 389
+LLLDEPTNHLDL A +WLEE L + + +VV+SH +DFLN VC+ IIH+ +L Y
Sbjct: 792 ILLLDEPTNHLDLEACVWLEESLKNFDRIMVVISHSQDFLNGVCTNIIHMQSKQLKMY 965
Score = 63.9 bits (154), Expect = 8e-11
Identities = 66/273 (24%), Positives = 108/273 (39%), Gaps = 26/273 (9%)
Frame = +3
Query: 427 KAQQDKVKDRAKFAAAKESKSKSKGKVDEDETQVEVPHKWRDYSVEFHFPEPTELTPPLL 486
KA Q K AK + + S D+ ++ + D + + PL
Sbjct: 114 KAAQKKAAAAAKRGGKAATAAASSKAAASDKAADKLANGIADMQISDRTCTGVLCSHPLS 293
Query: 487 QLIEV-SFSYPNREDFRLSDVDVGIDMGTRVAIVGPNGAGKSTLLNLL------------ 533
+ I + S S + D ++ ++ G R ++G NG GKSTLL +
Sbjct: 294 RDIRIESLSVTFHGHDLIVDSELELNYGRRYGLLGLNGCGKSTLLTAIGHRELPIPEHMD 473
Query: 534 ---------AGDLVPSEGEVR-RSQKLRIGRYSQHFVDLLTMDETPVQYLLRLHPDQEGL 583
A D+ E V ++LR+ + ++ L D+ + L R++ E L
Sbjct: 474 IYHLTREIEASDMSALEAVVSCDEERLRLEKEAE---TLAAQDDGGGETLERIYERLEAL 644
Query: 584 SKQEAVRAK---LGKYGLPSHNHLTPIVKLSGGQKARVVFTSISMSRPHILLLDEPTNHL 640
A + L G SGG + R+ P ILLLDEPTNHL
Sbjct: 645 DASTAEKRAAEILHGLGFDKKMQAKKTRDFSGGWRMRIALARALFINPTILLLDEPTNHL 824
Query: 641 DMQSIDALADALDEFTGGVVLVSHDSRLISRVC 673
D+++ L ++L F +V++SH ++ VC
Sbjct: 825 DLEACVWLEESLKNFDRIMVVISHSQDFLNGVC 923
>TC9119 homologue to AAQ65167 (AAQ65167) At1g64550, partial (20%)
Length = 666
Score = 130 bits (326), Expect = 9e-31
Identities = 66/149 (44%), Positives = 98/149 (65%)
Frame = +3
Query: 555 YSQHFVDLLTMDETPVQYLLRLHPDQEGLSKQEAVRAKLGKYGLPSHNHLTPIVKLSGGQ 614
+SQH VD L + P+ Y++R +P G+ +Q+ +RA LG +G+ + L P+ LSGGQ
Sbjct: 6 FSQHHVDGLDLSSNPLLYMMRCYP---GVPEQK-LRAHLGSFGVTGNLALQPMYTLSGGQ 173
Query: 615 KARVVFTSISMSRPHILLLDEPTNHLDMQSIDALADALDEFTGGVVLVSHDSRLISRVCD 674
K+RV F I+ +PHI+LLDEP++HLD+ +++AL L F GG+++VSHD LIS D
Sbjct: 174 KSRVAFAKITFKKPHIILLDEPSHHLDLDAVEALIQGLVMFQGGILMVSHDEHLISGSVD 353
Query: 675 DEERSQIWVVEDGTVRNFPGTFEDYKEDL 703
++WVV GT F G+F+DYK+ L
Sbjct: 354 -----ELWVVSQGTATPFHGSFQDYKKIL 425
Score = 57.8 bits (138), Expect = 6e-09
Identities = 29/84 (34%), Positives = 51/84 (60%)
Frame = +3
Query: 295 LAGLGFTKDMQGRPTKSFSGGWRMRISLARALFVQPTLLLLDEPTNHLDLRAVLWLEEYL 354
L G T ++ +P + SGG + R++ A+ F +P ++LLDEP++HLDL AV L + L
Sbjct: 108 LGSFGVTGNLALQPMYTLSGGQKSRVAFAKITFKKPHIILLDEPSHHLDLDAVEALIQGL 287
Query: 355 CRWKKTLVVVSHDRDFLNTVCSEI 378
++ +++VSHD ++ E+
Sbjct: 288 VMFQGGILMVSHDEHLISGSVDEL 359
>AV410532
Length = 418
Score = 117 bits (293), Expect = 6e-27
Identities = 57/83 (68%), Positives = 65/83 (77%)
Frame = +1
Query: 1 MGKKKSEDAGPSTKAKPGSKDVSKKEKFSVSAMLAGMDEKADKPKKASSNKAKPKPAPKA 60
MGKKK++DAG STKAK SKD KKEK SVSAMLA MDEK DK KK +S+ KPKP PKA
Sbjct: 163 MGKKKTDDAGQSTKAKASSKDAPKKEKISVSAMLASMDEKPDKAKKVASSSNKPKPKPKA 342
Query: 61 SAYTDDIDLPPSDDDESEEEQEE 83
S YTD IDLPPSDDD+ + ++E
Sbjct: 343 STYTDGIDLPPSDDDDDDLSEDE 411
>TC17035 similar to UP|Q9M1H3 (Q9M1H3) ABC transporter-like protein
(At3g54540), partial (6%)
Length = 590
Score = 94.7 bits (234), Expect = 4e-20
Identities = 43/45 (95%), Positives = 45/45 (99%)
Frame = +1
Query: 669 ISRVCDDEERSQIWVVEDGTVRNFPGTFEDYKEDLLKEIKAEVDD 713
ISRVCDDEERSQIW+VEDGTVRNFPGTFEDYK+DLLKEIKAEVDD
Sbjct: 1 ISRVCDDEERSQIWIVEDGTVRNFPGTFEDYKDDLLKEIKAEVDD 135
>BP055390
Length = 488
Score = 47.4 bits (111), Expect = 8e-06
Identities = 35/84 (41%), Positives = 48/84 (56%), Gaps = 2/84 (2%)
Frame = +3
Query: 608 VKLSGGQKARVVFTSISMSRPHILLLDEPTNHLDMQSIDALADALDEFTGG--VVLVSHD 665
V+LSGGQK R+ + P ILLLDE T+ LD +S + +AL+ G V+L++H
Sbjct: 225 VQLSGGQKQRIAIARAILKNPPILLLDEATSALDTESEKLVQEALETAMHGRTVILIAH- 401
Query: 666 SRLISRVCDDEERSQIWVVEDGTV 689
RL + V D I VVE+G V
Sbjct: 402 -RLSTVVNAD----VIAVVENGQV 458
Score = 41.6 bits (96), Expect = 4e-04
Identities = 22/63 (34%), Positives = 38/63 (59%), Gaps = 2/63 (3%)
Frame = +3
Query: 306 GRPTKSFSGGWRMRISLARALFVQPTLLLLDEPTNHLDLRAVLWLEEYL--CRWKKTLVV 363
G+ SGG + RI++ARA+ P +LLLDE T+ LD + ++E L +T+++
Sbjct: 213 GQRGVQLSGGQKQRIAIARAILKNPPILLLDEATSALDTESEKLVQEALETAMHGRTVIL 392
Query: 364 VSH 366
++H
Sbjct: 393 IAH 401
>TC15722 similar to UP|Q9FMU0 (Q9FMU0) Similarity to ABC transporter,
partial (37%)
Length = 624
Score = 47.0 bits (110), Expect = 1e-05
Identities = 34/83 (40%), Positives = 49/83 (58%), Gaps = 3/83 (3%)
Frame = +3
Query: 295 LAGLGFTKDMQGRPTKSFSGGWRMRISLARALFVQPTLLLLDEPTNHLDLRAVLWLEEYL 354
L+G+ K+ Q S SGG++ R++LA L P LL+LDEP LD +A + + L
Sbjct: 33 LSGISLDKNPQ-----SLSGGYKRRLALAIQLVQVPDLLILDEPLAGLDWKARADVVKLL 197
Query: 355 CRWKK--TLVVVSHD-RDFLNTV 374
KK T++VVSHD R+F + V
Sbjct: 198 KHLKKELTVLVVSHDLREFASLV 266
Score = 35.0 bits (79), Expect = 0.041
Identities = 25/76 (32%), Positives = 38/76 (49%), Gaps = 4/76 (5%)
Frame = +3
Query: 610 LSGGQKARVVFTSISMSRPHILLLDEPTNHLDMQS----IDALADALDEFTGGVVLVSHD 665
LSGG K R+ + P +L+LDEP LD ++ + L E T V++VSHD
Sbjct: 69 LSGGYKRRLALAIQLVQVPDLLILDEPLAGLDWKARADVVKLLKHLKKELT--VLVVSHD 242
Query: 666 SRLISRVCDDEERSQI 681
R + + D R ++
Sbjct: 243 LREFASLVDRSWRMEM 290
>TC11425 similar to UP|Q9M1Q9 (Q9M1Q9) P-glycoprotein-like proetin, partial
(9%)
Length = 712
Score = 45.4 bits (106), Expect = 3e-05
Identities = 29/81 (35%), Positives = 47/81 (57%), Gaps = 1/81 (1%)
Frame = +2
Query: 610 LSGGQKARVVFTSISMSRPHILLLDEPTNHLDMQSIDALADALDEFTGGVVLVSHDSRLI 669
LSGGQK RV + P+ILLLDE T+ LD +S + DALD+ V+V+ + ++
Sbjct: 107 LSGGQKQRVAIARAIIKTPNILLLDEATSALDAESERVVQDALDK-----VMVNRTTVIV 271
Query: 670 S-RVCDDEERSQIWVVEDGTV 689
+ R+ + I V+++G +
Sbjct: 272 AHRLSTIKNADVITVLKNGVI 334
Score = 40.4 bits (93), Expect = 0.001
Identities = 21/56 (37%), Positives = 36/56 (63%), Gaps = 2/56 (3%)
Frame = +2
Query: 313 SGGWRMRISLARALFVQPTLLLLDEPTNHLDLRAVLWLEEYLCR--WKKTLVVVSH 366
SGG + R+++ARA+ P +LLLDE T+ LD + +++ L + +T V+V+H
Sbjct: 110 SGGQKQRVAIARAIIKTPNILLLDEATSALDAESERVVQDALDKVMVNRTTVIVAH 277
>BG662257
Length = 336
Score = 45.1 bits (105), Expect = 4e-05
Identities = 23/46 (50%), Positives = 29/46 (63%)
Frame = +1
Query: 608 VKLSGGQKARVVFTSISMSRPHILLLDEPTNHLDMQSIDALADALD 653
++LSGGQK RV + P ILLLDE T+ LD +S + DALD
Sbjct: 133 IQLSGGQKQRVAIARAIVKSPKILLLDEATSALDAESEKVVQDALD 270
Score = 42.0 bits (97), Expect = 3e-04
Identities = 22/56 (39%), Positives = 37/56 (65%), Gaps = 2/56 (3%)
Frame = +1
Query: 313 SGGWRMRISLARALFVQPTLLLLDEPTNHLDLRAVLWLEEYLCRWK--KTLVVVSH 366
SGG + R+++ARA+ P +LLLDE T+ LD + +++ L R + +T +VV+H
Sbjct: 142 SGGQKQRVAIARAIVKSPKILLLDEATSALDAESEKVVQDALDRVRVDRTTIVVAH 309
>AW720269
Length = 524
Score = 44.3 bits (103), Expect = 7e-05
Identities = 45/143 (31%), Positives = 72/143 (49%), Gaps = 18/143 (12%)
Frame = +3
Query: 516 VAIVGPNGAGKSTLLNLLAG-----DLVP-----SEGEVRRSQKLR--IGRYSQ--HFVD 561
VA+VGP+G GKSTLL ++AG D P ++ + +LR G +Q + +
Sbjct: 96 VAVVGPSGTGKSTLLRIIAGRVKDRDFDPKTISINDHPMTSPAQLRKICGFVAQEDNLLP 275
Query: 562 LLTMDET---PVQYLLRLHPDQEGLSKQEAVRAKLGKYGL-PSHNHLTPIVKLSGGQKAR 617
LLT+ ET ++ L+ + + E++ +LG + + S +SGG++ R
Sbjct: 276 LLTVKETLLFSAKFRLKEMTPNDREMRVESLMQELGLFHVSDSFVGDEENRGISGGERKR 455
Query: 618 VVFTSISMSRPHILLLDEPTNHL 640
V + P ILLLDEPT+ L
Sbjct: 456 VSIGVDMIHNPPILLLDEPTSGL 524
Score = 41.6 bits (96), Expect = 4e-04
Identities = 46/178 (25%), Positives = 77/178 (42%), Gaps = 3/178 (1%)
Frame = +3
Query: 168 LLKNTSVKISHGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQEVVGDDKTAL 227
+LK+ S + ++GP+G GKSTLL+++A R K+ D D KT
Sbjct: 54 ILKSVSFVARSSEIVAVVGPSGTGKSTLLRIIAGRV----KDRDF---------DPKT-- 188
Query: 228 EAVVSANVELIKVRQKVADLQNIASGEEGMDKDDTNEEE---DAGEKLAELYEQLQLMGS 284
+S N + ++ + + E+ + T +E A +L E+ + M
Sbjct: 189 ---ISINDHPMTSPAQLRKICGFVAQEDNLLPLLTVKETLLFSAKFRLKEMTPNDREMRV 359
Query: 285 DAAESQASKILAGLGFTKDMQGRPTKSFSGGWRMRISLARALFVQPTLLLLDEPTNHL 342
++ + F D + R SGG R R+S+ + P +LLLDEPT+ L
Sbjct: 360 ESLMQELGLFHVSDSFVGDEENR---GISGGERKRVSIGVDMIHNPPILLLDEPTSGL 524
>TC10604 similar to AAQ89671 (AAQ89671) At4g33460, partial (36%)
Length = 498
Score = 43.9 bits (102), Expect = 9e-05
Identities = 30/80 (37%), Positives = 43/80 (53%), Gaps = 2/80 (2%)
Frame = +2
Query: 489 IEVSFSYPNREDFR-LSDVDVGIDMGTRVAIVGPNGAGKSTLLNLLAGDLVPSEGEVRRS 547
+ SF+ D R L D + I G ++GPNG GKSTLL +LAG L P+ G V +
Sbjct: 158 LNFSFTARQTNDVRVLRDCSLRIPSGQFWMLLGPNGCGKSTLLKILAGLLAPTSGTVYVN 337
Query: 548 Q-KLRIGRYSQHFVDLLTMD 566
+ K + + H V + T+D
Sbjct: 338 EPKSFVFQNPDHQVVMPTVD 397
Score = 43.9 bits (102), Expect = 9e-05
Identities = 30/87 (34%), Positives = 47/87 (53%), Gaps = 7/87 (8%)
Frame = +2
Query: 133 VIGSRASVLDGDDGADANVKDITIE----NFSVAARGKE---LLKNTSVKISHGKRYGLI 185
++ R S L + +V+++ I NFS AR +L++ S++I G+ + L+
Sbjct: 74 LLAPRCSTLPTNTIRSNSVENVAIVGRNLNFSFTARQTNDVRVLRDCSLRIPSGQFWMLL 253
Query: 186 GPNGMGKSTLLKLLAWRKIPVPKNIDV 212
GPNG GKSTLLK+LA P + V
Sbjct: 254 GPNGCGKSTLLKILAGLLAPTSGTVYV 334
>BI417718
Length = 515
Score = 42.4 bits (98), Expect = 3e-04
Identities = 20/45 (44%), Positives = 31/45 (68%), Gaps = 1/45 (2%)
Frame = +2
Query: 486 LQLIEVSFSYPNREDFRL-SDVDVGIDMGTRVAIVGPNGAGKSTL 529
++ VSF YP+R D ++ D+ + I GT VA+VG +G+GKST+
Sbjct: 380 IEFCHVSFKYPSRPDIQIFPDLSLTIHAGTTVALVGESGSGKSTV 514
>AV776921
Length = 435
Score = 42.0 bits (97), Expect = 3e-04
Identities = 42/156 (26%), Positives = 78/156 (49%), Gaps = 27/156 (17%)
Frame = +2
Query: 490 EVSFSYPNREDFR-LSDVDVGIDMGTRVAIVGPNGAGKSTLLNL-------LAGDLVPSE 541
+V FSYP+R D L+ + + I G VA+VG +G+GKST+++L L+GD++
Sbjct: 2 DVCFSYPSRPDVEILNKLCLDIPSGKIVALVGGSGSGKSTVISLIERFYEPLSGDILLDG 181
Query: 542 GEVR----RSQKLRIGRYSQ--------------HFVDLLTMDETPVQYLLRLHPDQEGL 583
++R + + +IG +Q + D T++E ++ ++L Q +
Sbjct: 182 NDIRDLDLKWLRQQIGLVNQEPALFATSIKENILYGKDNATLEE--LKRAVKLSDAQSFI 355
Query: 584 SK-QEAVRAKLGKYGLPSHNHLTPIVKLSGGQKARV 618
+ E + ++G+ G ++LSGGQK R+
Sbjct: 356 NNLPERLETQVGERG----------IQLSGGQKQRI 433
Score = 30.4 bits (67), Expect = 1.0
Identities = 17/48 (35%), Positives = 27/48 (55%)
Frame = +2
Query: 167 ELLKNTSVKISHGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLL 214
E+L + I GK L+G +G GKST++ L+ + P + D+LL
Sbjct: 38 EILNKLCLDIPSGKIVALVGGSGSGKSTVISLI--ERFYEPLSGDILL 175
>TC18647 similar to UP|Q9ASR9 (Q9ASR9) At2g01320/F10A8.20, partial (17%)
Length = 430
Score = 42.0 bits (97), Expect = 3e-04
Identities = 23/65 (35%), Positives = 39/65 (59%), Gaps = 4/65 (6%)
Frame = +1
Query: 480 ELTPPLLQLIEVSFSYPNRED----FRLSDVDVGIDMGTRVAIVGPNGAGKSTLLNLLAG 535
++TP ++ ++ S+ ++ F L +V G +AI+GP+G+GK+TLLN+LAG
Sbjct: 28 KVTPVTIRWSNINCSFSDKSSKSVRFLLKNVSGEAKPGRLLAIMGPSGSGKTTLLNVLAG 207
Query: 536 DLVPS 540
L S
Sbjct: 208 QLAAS 222
Score = 37.7 bits (86), Expect = 0.006
Identities = 24/75 (32%), Positives = 38/75 (50%), Gaps = 8/75 (10%)
Frame = +1
Query: 142 DGDDGADANVKDITIE----NFSVAARGKE----LLKNTSVKISHGKRYGLIGPNGMGKS 193
DG+ V +TI N S + + + LLKN S + G+ ++GP+G GK+
Sbjct: 4 DGETSPSGKVTPVTIRWSNINCSFSDKSSKSVRFLLKNVSGEAKPGRLLAIMGPSGSGKT 183
Query: 194 TLLKLLAWRKIPVPK 208
TLL +LA + P+
Sbjct: 184 TLLNVLAGQLAASPR 228
>AV417384
Length = 380
Score = 38.9 bits (89), Expect = 0.003
Identities = 19/42 (45%), Positives = 26/42 (61%)
Frame = +2
Query: 503 LSDVDVGIDMGTRVAIVGPNGAGKSTLLNLLAGDLVPSEGEV 544
L V+V + G + + G NG+GKST L +LAG PS GE+
Sbjct: 248 LRHVNVSLHDGGALVLTGANGSGKSTFLRMLAGFSRPSAGEI 373
>TC9999 similar to UP|Q9FMU0 (Q9FMU0) Similarity to ABC transporter,
partial (43%)
Length = 656
Score = 38.1 bits (87), Expect = 0.005
Identities = 33/119 (27%), Positives = 53/119 (43%), Gaps = 11/119 (9%)
Frame = +3
Query: 476 PEPTELTPPL-----------LQLIEVSFSYPNREDFRLSDVDVGIDMGTRVAIVGPNGA 524
P P + PPL ++ +VS+ P + L+ V + + I G +G+
Sbjct: 204 PPPFKFPPPLPLLRINCAHSSFEVRDVSYQAPGTQLKLLNSVTFSLPEKSFGLIFGQSGS 383
Query: 525 GKSTLLNLLAGDLVPSEGEVRRSQKLRIGRYSQHFVDLLTMDETPVQYLLRLHPDQEGL 583
GK+TLL LLAG P+ G + Y Q++ D D +P Q L P++ G+
Sbjct: 384 GKTTLLQLLAGINKPTSGSI----------YIQNYGD----DGSPSQSQEPLVPERVGI 518
Score = 29.6 bits (65), Expect = 1.7
Identities = 23/57 (40%), Positives = 31/57 (54%), Gaps = 2/57 (3%)
Frame = +3
Query: 156 IENFSVAARGKELLKNTSVKIS-HGKRYGLI-GPNGMGKSTLLKLLAWRKIPVPKNI 210
+ + S A G +L SV S K +GLI G +G GK+TLL+LLA P +I
Sbjct: 273 VRDVSYQAPGTQLKLLNSVTFSLPEKSFGLIFGQSGSGKTTLLQLLAGINKPTSGSI 443
>TC14999 similar to UP|Q9FJH6 (Q9FJH6) ABC transporter homolog PnATH-like
protein (At5g60790), partial (9%)
Length = 592
Score = 37.7 bits (86), Expect = 0.006
Identities = 23/60 (38%), Positives = 35/60 (58%)
Frame = +2
Query: 654 EFTGGVVLVSHDSRLISRVCDDEERSQIWVVEDGTVRNFPGTFEDYKEDLLKEIKAEVDD 713
E+ G ++LVSHD RLI++V +IWV E+ V+ + G D+K L + KA + D
Sbjct: 2 EWDGVLLLVSHDFRLINQVA-----HEIWVCENQAVKKWEGGIMDFKRHL--KAKAGLSD 160
>CB827630
Length = 405
Score = 37.4 bits (85), Expect = 0.008
Identities = 18/69 (26%), Positives = 36/69 (52%), Gaps = 2/69 (2%)
Frame = +2
Query: 312 FSGGWRMRISLARALFVQPTLLLLDEPTNHLD--LRAVLWLEEYLCRWKKTLVVVSHDRD 369
+SGG + R+ +A +L P ++ +DEP+ LD R LW L + + +++ +H +
Sbjct: 137 YSGGMKRRLIVAISLIGDPRVVYMDEPSTGLDPASRKSLWNVVKLAKRDRAIILTTHSME 316
Query: 370 FLNTVCSEI 378
+C +
Sbjct: 317 EAEALCDRL 343
Score = 34.3 bits (77), Expect = 0.071
Identities = 19/68 (27%), Positives = 33/68 (47%), Gaps = 2/68 (2%)
Frame = +2
Query: 609 KLSGGQKARVVFTSISMSRPHILLLDEPTNHLDMQSIDALADA--LDEFTGGVVLVSHDS 666
K SGG K R++ + P ++ +DEP+ LD S +L + L + ++L +H
Sbjct: 134 KYSGGMKRRLIVAISLIGDPRVVYMDEPSTGLDPASRKSLWNVVKLAKRDRAIILTTHSM 313
Query: 667 RLISRVCD 674
+CD
Sbjct: 314 EEAEALCD 337
>TC8582 similar to UP|Q8LJS2 (Q8LJS2) Nucleolar histone deacetylase
HD2-P39, partial (65%)
Length = 1341
Score = 37.0 bits (84), Expect = 0.011
Identities = 29/118 (24%), Positives = 48/118 (40%), Gaps = 1/118 (0%)
Frame = +1
Query: 5 KSEDAGPSTKAKPGSKDVSKKEKFSVSAMLAGMDEKADKPKKASSNKAKPKPAPKASAYT 64
+ E+ P TK + V+K K + +A + K PKK +P S Y
Sbjct: 418 EQENGKPETKTEDAK--VAKTAKPAAAAGPSAKQVKIVDPKKDEEEADLDVSSPDVSGYE 591
Query: 65 DD-IDLPPSDDDESEEEQEEKHRPDLKPLEVSIAEKELKKREKKDILAAHVAEQAKKE 121
DD I DDES++E +++ +V +K + K ++ A+ A E
Sbjct: 592 DDLISADEDSDDESDDESDDEEETPTPAKKVDQGKKRPNESASKTPVSGKKAKNATPE 765
>AV416268
Length = 424
Score = 37.0 bits (84), Expect = 0.011
Identities = 20/56 (35%), Positives = 30/56 (52%)
Frame = +3
Query: 586 QEAVRAKLGKYGLPSHNHLTPIVKLSGGQKARVVFTSISMSRPHILLLDEPTNHLD 641
Q+A+ ++G +G +SGGQK RV ++ P +L LDEPT+ LD
Sbjct: 123 QDAINTRIGGWGSKG---------VSGGQKRRVSICIEILTHPRLLFLDEPTSGLD 263
Score = 37.0 bits (84), Expect = 0.011
Identities = 18/41 (43%), Positives = 24/41 (57%)
Frame = +3
Query: 306 GRPTKSFSGGWRMRISLARALFVQPTLLLLDEPTNHLDLRA 346
G +K SGG + R+S+ + P LL LDEPT+ LD A
Sbjct: 150 GWGSKGVSGGQKRRVSICIEILTHPRLLFLDEPTSGLDSAA 272
>TC11559 similar to UP|Q9SXT3 (Q9SXT3) Multidrug resistance protein
(Fragment), partial (41%)
Length = 460
Score = 37.0 bits (84), Expect = 0.011
Identities = 30/111 (27%), Positives = 54/111 (48%), Gaps = 1/111 (0%)
Frame = +3
Query: 165 GKELLKNTSVKISHGKRYGLIGPNGMGKSTLLKLLAWRKIPVPKNIDVLLVEQEVVGDDK 224
G +LK + I G G+IGP+G GKSTLL+ L ++ P + V L +++
Sbjct: 126 GVPILKGIHLDIPKGVIVGVIGPSGSGKSTLLRAL--NRLWEPPSASVFLDARDI----- 284
Query: 225 TALEAVVSANVELIKVRQKVADLQNIASGEEGMDKDDTN-EEEDAGEKLAE 274
+++++ +R+KV L + + EG D+ + G+KL +
Sbjct: 285 --------CHLDVLSLRRKVGMLFQLPALFEGTVADNVRYGPQLRGQKLTD 413
Score = 32.3 bits (72), Expect = 0.27
Identities = 14/31 (45%), Positives = 20/31 (64%)
Frame = +3
Query: 503 LSDVDVGIDMGTRVAIVGPNGAGKSTLLNLL 533
L + + I G V ++GP+G+GKSTLL L
Sbjct: 138 LKGIHLDIPKGVIVGVIGPSGSGKSTLLRAL 230
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.313 0.133 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,252,329
Number of Sequences: 28460
Number of extensions: 138750
Number of successful extensions: 900
Number of sequences better than 10.0: 87
Number of HSP's better than 10.0 without gapping: 804
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 876
length of query: 713
length of database: 4,897,600
effective HSP length: 97
effective length of query: 616
effective length of database: 2,136,980
effective search space: 1316379680
effective search space used: 1316379680
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC136504.2


