

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136504.11 + phase: 0
(137 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
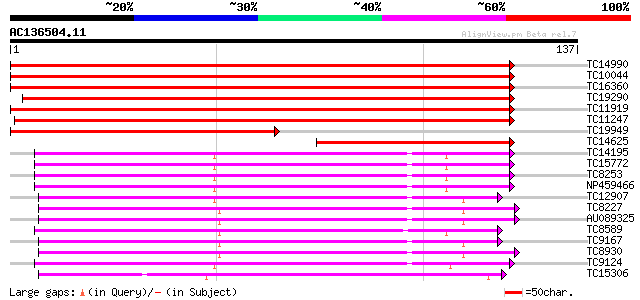
Score E
Sequences producing significant alignments: (bits) Value
TC14990 UP|Q8S2V3 (Q8S2V3) Small G-protein ROP9, complete 243 8e-66
TC10044 UP|O04369 (O04369) RAC1, complete 241 4e-65
TC16360 homologue to UP|Q9M628 (Q9M628) Small GTP binding protei... 236 8e-64
TC19290 homologue to UP|Q9SXT7 (Q9SXT7) Rac-type small GTP-bindi... 235 2e-63
TC11919 UP|RAC2_LOTJA (Q40220) RAC-like GTP binding protein RAC2... 232 1e-62
TC11247 homologue to UP|O82482 (O82482) RAC GTP binding protein ... 223 9e-60
TC19949 UP|Q8H0D4 (Q8H0D4) Rac GTPase, partial (33%) 135 2e-33
TC14625 homologue to UP|RAC1_PEA (Q35638) RAC-like GTP binding p... 96 2e-21
TC14195 UP|Q40215 (Q40215) RAB8A, complete 83 2e-17
TC15772 UP|Q40218 (Q40218) RAB8D, complete 82 3e-17
TC8253 UP|Q40219 (Q40219) RAB8E, complete 82 4e-17
NP459466 RAB8C [Lotus japonicus] 80 2e-16
TC12907 UP|Q40201 (Q40201) RAB1A, complete 80 2e-16
TC8227 UP|Q40203 (Q40203) RAB1C, complete 79 2e-16
AU089325 78 6e-16
TC8589 UP|Q40210 (Q40210) RAB5B, complete 77 8e-16
TC9167 UP|Q40202 (Q40202) RAB1B (Fragment), complete 77 8e-16
TC8930 UP|Q40204 (Q40204) RAB1D, complete 77 1e-15
TC9124 homologue to UP|Q41024 (Q41024) Small GTP-binding protein... 76 2e-15
TC15306 UP|Q40207 (Q40207) RAB1Y (Fragment), complete 75 3e-15
>TC14990 UP|Q8S2V3 (Q8S2V3) Small G-protein ROP9, complete
Length = 1029
Score = 243 bits (620), Expect = 8e-66
Identities = 119/122 (97%), Positives = 120/122 (97%)
Frame = +3
Query: 1 MSASRFIKCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVVVNGSTVNLGLWD 60
MSASRFIKCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVVVNGS VNLGLWD
Sbjct: 228 MSASRFIKCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVVVNGSIVNLGLWD 407
Query: 61 TAGQEDYNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPELKHYAPGVPIILVGTKL 120
TAGQEDYNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPELKHYAPGVPIILVGTKL
Sbjct: 408 TAGQEDYNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPELKHYAPGVPIILVGTKL 587
Query: 121 GM 122
+
Sbjct: 588 DL 593
Score = 24.3 bits (51), Expect = 8.1
Identities = 14/43 (32%), Positives = 20/43 (45%)
Frame = -3
Query: 11 TVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVVVNGST 53
TVG +VGK L+ + PT PTV + ++ ST
Sbjct: 343 TVGT*SVGKVLLV*EISKQVLPTAPSPTVTHLMNLEALIFSST 215
>TC10044 UP|O04369 (O04369) RAC1, complete
Length = 1292
Score = 241 bits (614), Expect = 4e-65
Identities = 116/122 (95%), Positives = 121/122 (99%)
Frame = +3
Query: 1 MSASRFIKCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVVVNGSTVNLGLWD 60
MSASRFIKCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVVV+GSTVNLGLWD
Sbjct: 213 MSASRFIKCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVVVDGSTVNLGLWD 392
Query: 61 TAGQEDYNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPELKHYAPGVPIILVGTKL 120
TAGQEDYNRLRPLSYRGADVFILAFSLISKASYEN++KKWIPEL+HYAPGVPIILVGTKL
Sbjct: 393 TAGQEDYNRLRPLSYRGADVFILAFSLISKASYENIAKKWIPELRHYAPGVPIILVGTKL 572
Query: 121 GM 122
+
Sbjct: 573 DL 578
>TC16360 homologue to UP|Q9M628 (Q9M628) Small GTP binding protein RACDP,
partial (64%)
Length = 479
Score = 236 bits (603), Expect = 8e-64
Identities = 112/122 (91%), Positives = 120/122 (97%)
Frame = +3
Query: 1 MSASRFIKCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVVVNGSTVNLGLWD 60
MSASRFIKCVTVGDGAVGKTC+LISYTSNTFPTDYVPTVFDNFSANVVV+GSTVNLGLWD
Sbjct: 102 MSASRFIKCVTVGDGAVGKTCMLISYTSNTFPTDYVPTVFDNFSANVVVDGSTVNLGLWD 281
Query: 61 TAGQEDYNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPELKHYAPGVPIILVGTKL 120
TAGQEDYNRLRPLSYRGADVF+L FSLIS+ASYENV+KKWIPEL+HYAPGVPIIL+GTKL
Sbjct: 282 TAGQEDYNRLRPLSYRGADVFLLCFSLISRASYENVAKKWIPELRHYAPGVPIILIGTKL 461
Query: 121 GM 122
+
Sbjct: 462 DL 467
>TC19290 homologue to UP|Q9SXT7 (Q9SXT7) Rac-type small GTP-binding protein,
partial (72%)
Length = 427
Score = 235 bits (600), Expect = 2e-63
Identities = 113/119 (94%), Positives = 118/119 (98%)
Frame = +3
Query: 4 SRFIKCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVVVNGSTVNLGLWDTAG 63
SRFIKCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVVV+GSTVNLGLWDTAG
Sbjct: 3 SRFIKCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVVVDGSTVNLGLWDTAG 182
Query: 64 QEDYNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPELKHYAPGVPIILVGTKLGM 122
QEDYNRLRPLSYRGADVF+LAFSLISKASYENV+KKWIPEL+HYAPGVPIILVGTKL +
Sbjct: 183 QEDYNRLRPLSYRGADVFLLAFSLISKASYENVAKKWIPELRHYAPGVPIILVGTKLDL 359
>TC11919 UP|RAC2_LOTJA (Q40220) RAC-like GTP binding protein RAC2, complete
Length = 974
Score = 232 bits (592), Expect = 1e-62
Identities = 109/122 (89%), Positives = 119/122 (97%)
Frame = +2
Query: 1 MSASRFIKCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVVVNGSTVNLGLWD 60
MS +RFIKCVTVGDGAVGKTC+LISYTSNTFPTDYVPTVFDNFSANVVV+GSTVNLGLWD
Sbjct: 113 MSTARFIKCVTVGDGAVGKTCMLISYTSNTFPTDYVPTVFDNFSANVVVDGSTVNLGLWD 292
Query: 61 TAGQEDYNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPELKHYAPGVPIILVGTKL 120
TAGQEDYNRLRPLSYRGADVF+LAFSL+S+ASYEN+SKKWIPEL+HYAP VPI+LVGTKL
Sbjct: 293 TAGQEDYNRLRPLSYRGADVFLLAFSLLSRASYENISKKWIPELRHYAPTVPIVLVGTKL 472
Query: 121 GM 122
+
Sbjct: 473 DL 478
>TC11247 homologue to UP|O82482 (O82482) RAC GTP binding protein ARAC8
(At3g48040), partial (60%)
Length = 559
Score = 223 bits (568), Expect = 9e-60
Identities = 103/121 (85%), Positives = 115/121 (94%)
Frame = +1
Query: 2 SASRFIKCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVVVNGSTVNLGLWDT 61
+ASRFIKCVTVGDGAVGKTC+LI YTSN FPTDY+PTVFDNFSANVV+ G TVNLGLWDT
Sbjct: 193 TASRFIKCVTVGDGAVGKTCMLICYTSNKFPTDYIPTVFDNFSANVVMEGITVNLGLWDT 372
Query: 62 AGQEDYNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPELKHYAPGVPIILVGTKLG 121
AGQEDYNRLRPLSYRGADVF+LAFSL+S+ASYENV KKWIPEL+H+APGVP++LVGTKL
Sbjct: 373 AGQEDYNRLRPLSYRGADVFVLAFSLVSRASYENVLKKWIPELQHFAPGVPVVLVGTKLD 552
Query: 122 M 122
+
Sbjct: 553 L 555
>TC19949 UP|Q8H0D4 (Q8H0D4) Rac GTPase, partial (33%)
Length = 549
Score = 135 bits (341), Expect = 2e-33
Identities = 65/65 (100%), Positives = 65/65 (100%)
Frame = +1
Query: 1 MSASRFIKCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVVVNGSTVNLGLWD 60
MSASRFIKCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVVVNGSTVNLGLWD
Sbjct: 355 MSASRFIKCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVVVNGSTVNLGLWD 534
Query: 61 TAGQE 65
TAGQE
Sbjct: 535 TAGQE 549
Score = 25.0 bits (53), Expect = 4.7
Identities = 13/29 (44%), Positives = 16/29 (54%)
Frame = -2
Query: 11 TVGDGAVGKTCLLISYTSNTFPTDYVPTV 39
TVG +VGK L+ + FPT PTV
Sbjct: 470 TVGT*SVGKVLLV*EISKQVFPTAPSPTV 384
>TC14625 homologue to UP|RAC1_PEA (Q35638) RAC-like GTP binding protein
RHO1, partial (62%)
Length = 709
Score = 95.9 bits (237), Expect = 2e-21
Identities = 46/48 (95%), Positives = 47/48 (97%)
Frame = +1
Query: 75 YRGADVFILAFSLISKASYENVSKKWIPELKHYAPGVPIILVGTKLGM 122
YRGADVFILAFSLISKASYENVSKKWIPELKHYAPGVPIILVGTKL +
Sbjct: 1 YRGADVFILAFSLISKASYENVSKKWIPELKHYAPGVPIILVGTKLDL 144
>TC14195 UP|Q40215 (Q40215) RAB8A, complete
Length = 1171
Score = 82.8 bits (203), Expect = 2e-17
Identities = 42/118 (35%), Positives = 69/118 (57%), Gaps = 2/118 (1%)
Frame = +2
Query: 7 IKCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVV-VNGSTVNLGLWDTAGQE 65
IK + +GD VGK+CLL+ ++ +F T ++ T+ +F + ++G + L +WDTAGQE
Sbjct: 248 IKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQE 427
Query: 66 DYNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPEL-KHYAPGVPIILVGTKLGM 122
+ + YRGA +L + + +AS+ N+ K WI + +H + V ILVG K M
Sbjct: 428 RFRTITTAYYRGAMGILLVYDVTDEASFNNI-KNWIRNIEQHASDNVNKILVGNKADM 598
>TC15772 UP|Q40218 (Q40218) RAB8D, complete
Length = 1139
Score = 82.0 bits (201), Expect = 3e-17
Identities = 42/118 (35%), Positives = 69/118 (57%), Gaps = 2/118 (1%)
Frame = +2
Query: 7 IKCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVV-VNGSTVNLGLWDTAGQE 65
IK + +GD VGK+CLL+ ++ +F T ++ T+ +F + ++G V L +WDTAGQE
Sbjct: 284 IKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRVKLQIWDTAGQE 463
Query: 66 DYNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPEL-KHYAPGVPIILVGTKLGM 122
+ + YRGA +L + + +AS+ N+ + WI + +H + V ILVG K M
Sbjct: 464 RFRTITTAYYRGAMGILLVYDVTDEASFNNI-RNWIRNIEQHASDNVNKILVGNKADM 634
>TC8253 UP|Q40219 (Q40219) RAB8E, complete
Length = 1106
Score = 81.6 bits (200), Expect = 4e-17
Identities = 41/118 (34%), Positives = 69/118 (57%), Gaps = 2/118 (1%)
Frame = +3
Query: 7 IKCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVV-VNGSTVNLGLWDTAGQE 65
IK + +GD VGK+CLL+ ++ +F T ++ T+ +F + ++G + L +WDTAGQE
Sbjct: 228 IKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQE 407
Query: 66 DYNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPEL-KHYAPGVPIILVGTKLGM 122
+ + YRGA +L + + ++S+ N+ K WI + +H + V ILVG K M
Sbjct: 408 RFRTITTAYYRGAMGILLVYDVTDESSFNNI-KNWIHNIEQHASDNVNKILVGNKADM 578
>NP459466 RAB8C [Lotus japonicus]
Length = 639
Score = 79.7 bits (195), Expect = 2e-16
Identities = 40/118 (33%), Positives = 68/118 (56%), Gaps = 2/118 (1%)
Frame = +1
Query: 7 IKCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVV-VNGSTVNLGLWDTAGQE 65
IK + +GD VGK+CLL+ ++ +F T ++ T+ +F + ++G + L +WDTAGQE
Sbjct: 46 IKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQE 225
Query: 66 DYNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPEL-KHYAPGVPIILVGTKLGM 122
+ YRGA +L + + ++S+ N+ + WI + +H + V ILVG K M
Sbjct: 226 RVRTITTAYYRGAKGILLVYDVTDESSFNNI-RNWIRNIEQHASDNVNKILVGNKADM 396
>TC12907 UP|Q40201 (Q40201) RAB1A, complete
Length = 869
Score = 79.7 bits (195), Expect = 2e-16
Identities = 41/114 (35%), Positives = 66/114 (56%), Gaps = 2/114 (1%)
Frame = +2
Query: 8 KCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVV-VNGSTVNLGLWDTAGQED 66
K + +GD +VGK+CLL+ + +++ Y+ T+ +F V + G T+ L +WDTAGQE
Sbjct: 140 KLLLIGDSSVGKSCLLLRFADDSYVDSYISTIGVDFKIRTVELQGKTIKLQIWDTAGQER 319
Query: 67 YNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPELKHYA-PGVPIILVGTK 119
+ + YRGA I+ + + S+ NV K+W+ E+ YA V +LVG K
Sbjct: 320 FRTITSSYYRGAHGIIIVYDVTEMESFNNV-KQWLNEIDRYANDSVCKLLVGNK 478
>TC8227 UP|Q40203 (Q40203) RAB1C, complete
Length = 1083
Score = 79.3 bits (194), Expect = 2e-16
Identities = 41/118 (34%), Positives = 68/118 (56%), Gaps = 2/118 (1%)
Frame = +2
Query: 8 KCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVVV-NGSTVNLGLWDTAGQED 66
K + +GD VGK+CLL+ + +++ Y+ T+ +F V +G T+ L +WDTAGQE
Sbjct: 155 KLLLIGDSGVGKSCLLLRFADDSYLDSYISTIGVDFKIRTVEQDGKTIKLQIWDTAGQER 334
Query: 67 YNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPELKHYA-PGVPIILVGTKLGMS 123
+ + YRGA I+ + + + S+ NV K+W+ E+ YA V +LVG K ++
Sbjct: 335 FRTITSSYYRGAHGIIVVYDVTDQESFNNV-KQWLNEIDRYASENVNKLLVGNKCDLT 505
>AU089325
Length = 757
Score = 77.8 bits (190), Expect = 6e-16
Identities = 40/118 (33%), Positives = 67/118 (55%), Gaps = 2/118 (1%)
Frame = +3
Query: 8 KCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVVV-NGSTVNLGLWDTAGQED 66
K + +GD VGK+CLL+ + +++ Y+ T+ +F V + T+ L +WDTAGQE
Sbjct: 75 KLLLIGDSGVGKSCLLLRFADDSYIESYISTIGVDFKIRTVEQDAKTIKLQIWDTAGQER 254
Query: 67 YNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPELKHYA-PGVPIILVGTKLGMS 123
+ + YRGA I+ + + + S+ NV K+W+ E+ YA V +LVG K ++
Sbjct: 255 FRTITSSYYRGAHGIIIVYDVTDEESFNNV-KQWLSEIDRYASDNVNKLLVGNKCDLT 425
>TC8589 UP|Q40210 (Q40210) RAB5B, complete
Length = 1241
Score = 77.4 bits (189), Expect = 8e-16
Identities = 42/116 (36%), Positives = 65/116 (55%), Gaps = 3/116 (2%)
Frame = +1
Query: 7 IKCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVVV--NGSTVNLGLWDTAGQ 64
+K V +GD VGK+C+++ + F TV +F + + + +TV +WDTAGQ
Sbjct: 322 VKLVLLGDSGVGKSCIVLRFVRGQFDPTSKVTVGASFLSQTIALQDSTTVKFEIWDTAGQ 501
Query: 65 EDYNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPEL-KHYAPGVPIILVGTK 119
E Y L PL YRGA V ++ + + S S+ ++ W+ EL KH P + + LVG K
Sbjct: 502 ERYAALAPLYYRGAAVAVIVYDITSPESFSK-AQYWVKELQKHGNPDIVMALVGNK 666
>TC9167 UP|Q40202 (Q40202) RAB1B (Fragment), complete
Length = 951
Score = 77.4 bits (189), Expect = 8e-16
Identities = 40/114 (35%), Positives = 65/114 (56%), Gaps = 2/114 (1%)
Frame = +2
Query: 8 KCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVVV-NGSTVNLGLWDTAGQED 66
K + +GD VGK+CLL+ + +++ Y+ T+ +F V + T+ L +WDTAGQE
Sbjct: 119 KLLLIGDSGVGKSCLLLRFADDSYIESYISTIGVDFKIRTVEQDAKTIKLQIWDTAGQER 298
Query: 67 YNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPELKHYA-PGVPIILVGTK 119
+ + YRGA I+ + + + S+ NV K+W+ E+ YA V +LVG K
Sbjct: 299 FRTITSSYYRGAHGIIIVYDVTDEESFNNV-KQWLSEIDRYASDNVNKLLVGNK 457
>TC8930 UP|Q40204 (Q40204) RAB1D, complete
Length = 943
Score = 77.0 bits (188), Expect = 1e-15
Identities = 40/118 (33%), Positives = 67/118 (55%), Gaps = 2/118 (1%)
Frame = +3
Query: 8 KCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVVV-NGSTVNLGLWDTAGQED 66
K + +GD VGK+CLL+ + +++ Y+ T+ +F V + T+ L +WDTAGQE
Sbjct: 117 KLLLIGDSGVGKSCLLLRFGDDSYIESYISTIGVDFKIRTVEQDAKTIKLQIWDTAGQER 296
Query: 67 YNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPELKHYA-PGVPIILVGTKLGMS 123
+ + YRGA I+ + + + S+ NV K+W+ E+ YA V +LVG K ++
Sbjct: 297 FRTITSSYYRGAHGIIIVYDVTDEESFNNV-KQWLSEIDRYASDNVNKLLVGNKCDLT 467
>TC9124 homologue to UP|Q41024 (Q41024) Small GTP-binding protein, complete
Length = 1109
Score = 76.3 bits (186), Expect = 2e-15
Identities = 39/118 (33%), Positives = 68/118 (57%), Gaps = 2/118 (1%)
Frame = +3
Query: 7 IKCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSANVV-VNGSTVNLGLWDTAGQE 65
IK + +GD VGK+CLL+ ++ +F T ++ T+ +F + ++G + L +WDTAGQE
Sbjct: 297 IKLLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLHIWDTAGQE 476
Query: 66 DYNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPELK-HYAPGVPIILVGTKLGM 122
+ + Y GA +L + + ++S+ ++ + WI L+ H + V ILVG K M
Sbjct: 477 RFRTITTAYYLGAMGILLVYDVTDESSFNHI-QNWIRHLEPHASDNVNKILVGNKADM 647
>TC15306 UP|Q40207 (Q40207) RAB1Y (Fragment), complete
Length = 1019
Score = 75.5 bits (184), Expect = 3e-15
Identities = 42/116 (36%), Positives = 65/116 (55%), Gaps = 3/116 (2%)
Frame = +2
Query: 8 KCVTVGDGAVGKTCLLISYTSNTFPTDYVPTVFDNFSAN-VVVNGSTVNLGLWDTAGQED 66
K + +GD VGK+ LL+S+TS+ F D PT+ +F V + G + L +WDTAGQE
Sbjct: 116 KLLMIGDSGVGKSSLLLSFTSDEFQ-DMSPTIGVDFKVKYVAIGGKKLKLAIWDTAGQER 292
Query: 67 YNRLRPLSYRGADVFILAFSLISKASYENVSKKWIPELKHYAPGVPII--LVGTKL 120
+ L YRGA I+ + + + ++ N+S+ W E+ Y+ I LVG K+
Sbjct: 293 FRTLTSSYYRGAQGIIMVYDVTRRDTFTNLSEIWAKEIDLYSTNQDCIKMLVGNKV 460
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.323 0.139 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,591,802
Number of Sequences: 28460
Number of extensions: 32171
Number of successful extensions: 253
Number of sequences better than 10.0: 76
Number of HSP's better than 10.0 without gapping: 217
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 217
length of query: 137
length of database: 4,897,600
effective HSP length: 81
effective length of query: 56
effective length of database: 2,592,340
effective search space: 145171040
effective search space used: 145171040
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 50 (23.9 bits)
Medicago: description of AC136504.11


