

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136472.17 - phase: 0
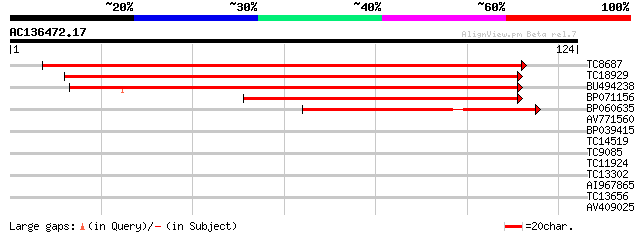
(124 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC8687 similar to UP|Q9SXL2 (Q9SXL2) Serine decarboxylase, parti... 198 3e-52
TC18929 125 2e-30
BU494238 117 6e-28
BP071156 83 2e-17
BP060635 54 6e-09
AV771560 28 0.60
BP039415 26 2.3
TC14519 25 3.0
TC9085 similar to UP|Q9M724 (Q9M724) Branched chain alpha-keto a... 25 3.9
TC11924 similar to UP|P93484 (P93484) BP-80 vacuolar sorting rec... 25 3.9
TC13302 24 6.6
AI967865 24 6.6
TC13656 24 8.7
AV409025 24 8.7
>TC8687 similar to UP|Q9SXL2 (Q9SXL2) Serine decarboxylase, partial (80%)
Length = 1644
Score = 198 bits (503), Expect = 3e-52
Identities = 92/106 (86%), Positives = 98/106 (91%)
Frame = +1
Query: 8 FEKEVQKCLRNAHYFKDRLIEAGIGAMLNELSSTVVFERPHDEEFIRKWQLACKGNIAHV 67
F+KEV KCLRNAHYFKDRL+EAGIGAMLNELSSTVVFERPHDEEFIRKWQLACKGNIAHV
Sbjct: 1192 FQKEVLKCLRNAHYFKDRLVEAGIGAMLNELSSTVVFERPHDEEFIRKWQLACKGNIAHV 1371
Query: 68 VVMPNVTIEKLDDFLNELVQKRATWFEYGTFQPYCIASDVGENSCL 113
VVMPN+TIEKLDDFL EL+Q RATWF+ G + PYCIASDVGE CL
Sbjct: 1372 VVMPNITIEKLDDFLEELMQNRATWFQDGKYTPYCIASDVGEKDCL 1509
>TC18929
Length = 563
Score = 125 bits (314), Expect = 2e-30
Identities = 57/100 (57%), Positives = 76/100 (76%)
Frame = +3
Query: 13 QKCLRNAHYFKDRLIEAGIGAMLNELSSTVVFERPHDEEFIRKWQLACKGNIAHVVVMPN 72
Q C+ NA + D+L + GIGAM NE S+TV+FERP D++F+RKW L+C+GN+AHVVVM +
Sbjct: 6 QICITNASHLLDKLHDNGIGAMRNEFSNTVIFERPLDDDFVRKWNLSCEGNVAHVVVMQH 185
Query: 73 VTIEKLDDFLNELVQKRATWFEYGTFQPYCIASDVGENSC 112
VTIE LD F++ELV KR WF+ G + CIA+ VG N+C
Sbjct: 186 VTIEMLDSFVSELVNKRKVWFQDGQRKSPCIANSVGGNNC 305
>BU494238
Length = 448
Score = 117 bits (293), Expect = 6e-28
Identities = 56/100 (56%), Positives = 73/100 (73%), Gaps = 1/100 (1%)
Frame = +2
Query: 14 KCLRNAHYFK-DRLIEAGIGAMLNELSSTVVFERPHDEEFIRKWQLACKGNIAHVVVMPN 72
KC H D+L + IGAM NE S+TVVFERP D++F+RKW L+C+GN+AHVVVM +
Sbjct: 20 KCASQKHVTSLDKLHDNDIGAMRNEFSNTVVFERPPDDDFVRKWNLSCEGNVAHVVVMQH 199
Query: 73 VTIEKLDDFLNELVQKRATWFEYGTFQPYCIASDVGENSC 112
VTIE LD F++ELV KR+ WF+ G +P CIA +VG +C
Sbjct: 200 VTIEMLDSFVSELVTKRSFWFQGGQRKPPCIADNVGGKNC 319
>BP071156
Length = 404
Score = 82.8 bits (203), Expect = 2e-17
Identities = 34/61 (55%), Positives = 48/61 (77%)
Frame = -3
Query: 52 FIRKWQLACKGNIAHVVVMPNVTIEKLDDFLNELVQKRATWFEYGTFQPYCIASDVGENS 111
F+RKW L+C+G++AHVVVM +VTIE +D F+ ELV+KR+ WF+ G +P CIA+ VG +
Sbjct: 402 FVRKWSLSCEGDVAHVVVMKHVTIEMIDSFVGELVKKRSIWFQGGQIKPPCIANTVGREN 223
Query: 112 C 112
C
Sbjct: 222 C 220
>BP060635
Length = 360
Score = 54.3 bits (129), Expect = 6e-09
Identities = 26/52 (50%), Positives = 36/52 (69%)
Frame = -3
Query: 65 AHVVVMPNVTIEKLDDFLNELVQKRATWFEYGTFQPYCIASDVGENSCLAYV 116
AHVVVM +VTIE +D F++ELV +R WF+ G P CIA+++G +C V
Sbjct: 358 AHVVVMQHVTIEMIDSFVSELVNQRTIWFQDGL--PPCIANNIGGENCACAV 209
>AV771560
Length = 445
Score = 27.7 bits (60), Expect = 0.60
Identities = 11/22 (50%), Positives = 16/22 (72%)
Frame = -3
Query: 74 TIEKLDDFLNELVQKRATWFEY 95
T KLDDF NE+V+ R +++ Y
Sbjct: 242 TSRKLDDFFNEIVEGRKSFWAY 177
>BP039415
Length = 449
Score = 25.8 bits (55), Expect = 2.3
Identities = 9/19 (47%), Positives = 13/19 (68%)
Frame = -3
Query: 49 DEEFIRKWQLACKGNIAHV 67
DE F R+W+L C+ AH+
Sbjct: 405 DESFSRRWKLRCQLQAAHL 349
>TC14519
Length = 1095
Score = 25.4 bits (54), Expect = 3.0
Identities = 11/25 (44%), Positives = 13/25 (52%)
Frame = +1
Query: 47 PHDEEFIRKWQLACKGNIAHVVVMP 71
P F R W+L C GN HV +P
Sbjct: 268 PRRSRFHRLWELHCTGNP*HVRGLP 342
>TC9085 similar to UP|Q9M724 (Q9M724) Branched chain alpha-keto acid
dehydrogenase E2 subunit, partial (45%)
Length = 912
Score = 25.0 bits (53), Expect = 3.9
Identities = 8/14 (57%), Positives = 12/14 (85%)
Frame = -3
Query: 91 TWFEYGTFQPYCIA 104
T+F +GT +PYC+A
Sbjct: 358 TFFMFGTTRPYCVA 317
>TC11924 similar to UP|P93484 (P93484) BP-80 vacuolar sorting receptor,
partial (41%)
Length = 783
Score = 25.0 bits (53), Expect = 3.9
Identities = 16/59 (27%), Positives = 28/59 (47%), Gaps = 6/59 (10%)
Frame = +3
Query: 61 KGNIAHVVVMPNVTIE------KLDDFLNELVQKRATWFEYGTFQPYCIASDVGENSCL 113
KG+ V ++P + + KL+ +++ + FE T C++SDV N CL
Sbjct: 42 KGSRGDVTILPTLVVNNRQYRGKLEK--GAVMKAICSGFEETTEPAVCLSSDVETNECL 212
>TC13302
Length = 533
Score = 24.3 bits (51), Expect = 6.6
Identities = 12/37 (32%), Positives = 20/37 (53%), Gaps = 2/37 (5%)
Frame = -2
Query: 67 VVVMPNVTIEKLDDF--LNELVQKRATWFEYGTFQPY 101
+V+ V++ +DD LN +V+K + YG PY
Sbjct: 466 MVIKGGVSLASVDDVITLNRVVEKPCIDYRYGENSPY 356
>AI967865
Length = 326
Score = 24.3 bits (51), Expect = 6.6
Identities = 13/58 (22%), Positives = 27/58 (46%), Gaps = 5/58 (8%)
Frame = +2
Query: 61 KGNIAHVVVMPNVTIEKLDDFLNEL-----VQKRATWFEYGTFQPYCIASDVGENSCL 113
+G+ V ++P + I + + +L ++ F+ T P C++ D+ N CL
Sbjct: 71 RGSRGDVTILPTLVINNVQ-YTGKLERTPVLKAVCAGFKESTEPPVCLSGDIETNECL 241
>TC13656
Length = 656
Score = 23.9 bits (50), Expect = 8.7
Identities = 7/16 (43%), Positives = 12/16 (74%)
Frame = -1
Query: 91 TWFEYGTFQPYCIASD 106
+WF+ T+Q C++SD
Sbjct: 239 SWFQKATYQVACVSSD 192
>AV409025
Length = 434
Score = 23.9 bits (50), Expect = 8.7
Identities = 10/34 (29%), Positives = 19/34 (55%)
Frame = -2
Query: 45 ERPHDEEFIRKWQLACKGNIAHVVVMPNVTIEKL 78
++PH+E R+ + C G++ PN T ++L
Sbjct: 178 KKPHEEREARE*R*ECDGSMERAKPSPNNTSDRL 77
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.324 0.138 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,403,143
Number of Sequences: 28460
Number of extensions: 27880
Number of successful extensions: 179
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 179
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 179
length of query: 124
length of database: 4,897,600
effective HSP length: 79
effective length of query: 45
effective length of database: 2,649,260
effective search space: 119216700
effective search space used: 119216700
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 50 (23.9 bits)
Medicago: description of AC136472.17


