

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136472.15 + phase: 0
(486 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
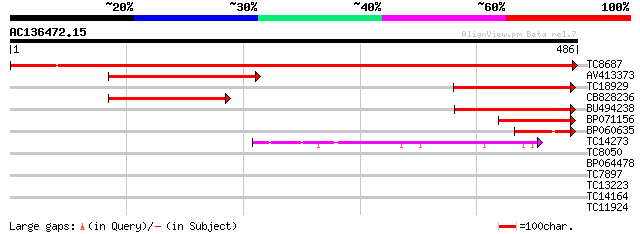
Score E
Sequences producing significant alignments: (bits) Value
TC8687 similar to UP|Q9SXL2 (Q9SXL2) Serine decarboxylase, parti... 897 0.0
AV413373 162 9e-41
TC18929 134 3e-32
CB828236 124 3e-29
BU494238 123 6e-29
BP071156 91 5e-19
BP060635 64 4e-11
TC14273 similar to UP|DCE1_ARATH (Q42521) Glutamate decarboxylas... 60 6e-10
TC8050 similar to UP|Q8LPQ5 (Q8LPQ5) AT3g18290/MIE15_8, partial ... 29 1.5
BP064478 28 3.4
TC7897 weakly similar to UP|ENL2_ARATH (Q9T076) Early nodulin-li... 28 4.4
TC13223 27 5.7
TC14164 weakly similar to UP|Q8VYH6 (Q8VYH6) AT4g17280/dl4675c, ... 27 9.8
TC11924 similar to UP|P93484 (P93484) BP-80 vacuolar sorting rec... 27 9.8
>TC8687 similar to UP|Q9SXL2 (Q9SXL2) Serine decarboxylase, partial (80%)
Length = 1644
Score = 897 bits (2319), Expect = 0.0
Identities = 433/486 (89%), Positives = 459/486 (94%)
Frame = +1
Query: 1 MVGSADVLVNGLSTNGAVELLPDDFDVSAIIKDPVPPVVAADNGIGKEEAKINGGKEKRE 60
MVGS VL + LS NGAVE LP+DFD +AIIKDPVPPVV +NG+ EA+++ GKEKRE
Sbjct: 70 MVGSFGVLADDLSINGAVEPLPEDFDTTAIIKDPVPPVVG-ENGVVNGEAQVSKGKEKRE 246
Query: 61 IVLGRNIHTTCLEVTEPEADDEITGDRDAHMASVLARYRKSLTERTKYHLGYPYNLDFDY 120
IVLGRNIHTTCLEVTEPEADDE+TGDR+A+MASVLARY+KSLTERTK+HLGYPYNLDFDY
Sbjct: 247 IVLGRNIHTTCLEVTEPEADDEVTGDREANMASVLARYKKSLTERTKHHLGYPYNLDFDY 426
Query: 121 GALSQLQHFSINNLGDPFIESNYGVHSRQFEVGVLDWFARLWELEKNEYWGYITNCGTEG 180
GALSQLQHFSINNLGDPFIESNYGVHSRQFEVGVLDWFARLWELEKNEYWGYITNCGTEG
Sbjct: 427 GALSQLQHFSINNLGDPFIESNYGVHSRQFEVGVLDWFARLWELEKNEYWGYITNCGTEG 606
Query: 181 NLHGILVGREVLPDGILYASRESHYSIFKAARMYRMECEKVETLNSGEIDCDDFKAKLLR 240
NLHGILVGREV PDG+LYASRESHYSIFKAARMYRMEC KV+TL SGEIDCDDFK KLL
Sbjct: 607 NLHGILVGREVYPDGVLYASRESHYSIFKAARMYRMECVKVDTLWSGEIDCDDFKTKLLL 786
Query: 241 HQDKPAIINVNIGTTVKGAVDDLDLVIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKRAP 300
HQDKPAIINVNIGTTVKGAVDDLD+VI+KLEEAGFS DRFYIHVDGALFGLMMPFVK+AP
Sbjct: 787 HQDKPAIINVNIGTTVKGAVDDLDMVIKKLEEAGFSHDRFYIHVDGALFGLMMPFVKQAP 966
Query: 301 KVTFKKPIGSVSVSGHKFVGCPMPCGVQITRLEHINALSRNVEYLASRDATIMGSRNGHA 360
KVTFKKPIGSVSVSGHKFVGCPMPCGVQITRLE++NALSR+VEYLASRDATIMGSRNGHA
Sbjct: 967 KVTFKKPIGSVSVSGHKFVGCPMPCGVQITRLEYVNALSRDVEYLASRDATIMGSRNGHA 1146
Query: 361 PIFLWYTLNRKGYRGFQKEVQKCLRNAHYFKDRLIEAGIGAMLNELSSTVVFERPHDEEF 420
PIFLWYTLNRKGYRGFQKEV KCLRNAHYFKDRL+EAGIGAMLNELSSTVVFERPHDEEF
Sbjct: 1147PIFLWYTLNRKGYRGFQKEVLKCLRNAHYFKDRLVEAGIGAMLNELSSTVVFERPHDEEF 1326
Query: 421 IRKWQLACKGNIAHVVVMPNVTIEKLDDFLNELVQKRATWFEDGTFQPYCIASDVGENSC 480
IRKWQLACKGNIAHVVVMPN+TIEKLDDFL EL+Q RATWF+DG + PYCIASDVGE C
Sbjct: 1327IRKWQLACKGNIAHVVVMPNITIEKLDDFLEELMQNRATWFQDGKYTPYCIASDVGEKDC 1506
Query: 481 LCAQHK 486
LCA HK
Sbjct: 1507LCALHK 1524
>AV413373
Length = 417
Score = 162 bits (411), Expect = 9e-41
Identities = 73/131 (55%), Positives = 94/131 (71%)
Frame = +1
Query: 85 GDRDAHMASVLARYRKSLTERTKYHLGYPYNLDFDYGALSQLQHFSINNLGDPFIESNYG 144
G+ A+++ ++A Y L + LG+P NLDFDY AL+ L + +NN GDPF+ S YG
Sbjct: 25 GETRANLSKIIANYVVMLNQNKSRCLGFPVNLDFDYDALTPLLQYHLNNAGDPFLGSGYG 204
Query: 145 VHSRQFEVGVLDWFARLWELEKNEYWGYITNCGTEGNLHGILVGREVLPDGILYASRESH 204
+S +FEV VLDWFA+LW++EK EYWGY+T GTEGNLHGIL+GRE +GILY S SH
Sbjct: 205 QNSAEFEVCVLDWFAKLWQMEKGEYWGYVTTGGTEGNLHGILMGREKFQNGILYTSENSH 384
Query: 205 YSIFKAARMYR 215
YS+FK MYR
Sbjct: 385 YSLFKIVMMYR 417
>TC18929
Length = 563
Score = 134 bits (337), Expect = 3e-32
Identities = 60/105 (57%), Positives = 79/105 (75%)
Frame = +3
Query: 381 QKCLRNAHYFKDRLIEAGIGAMLNELSSTVVFERPHDEEFIRKWQLACKGNIAHVVVMPN 440
Q C+ NA + D+L + GIGAM NE S+TV+FERP D++F+RKW L+C+GN+AHVVVM +
Sbjct: 6 QICITNASHLLDKLHDNGIGAMRNEFSNTVIFERPLDDDFVRKWNLSCEGNVAHVVVMQH 185
Query: 441 VTIEKLDDFLNELVQKRATWFEDGTFQPYCIASDVGENSCLCAQH 485
VTIE LD F++ELV KR WF+DG + CIA+ VG N+C C H
Sbjct: 186 VTIEMLDSFVSELVNKRKVWFQDGQRKSPCIANSVGGNNCACGLH 320
>CB828236
Length = 552
Score = 124 bits (311), Expect = 3e-29
Identities = 53/105 (50%), Positives = 75/105 (70%)
Frame = +3
Query: 85 GDRDAHMASVLARYRKSLTERTKYHLGYPYNLDFDYGALSQLQHFSINNLGDPFIESNYG 144
G+ +++ +A+Y L + LG+P N +F+Y AL+ L H+ +NN DPF+ S YG
Sbjct: 237 GESRTNLSKTIAKYVVKLNQNKSQCLGFPVNQEFNYDALTPLLHYHLNNASDPFLGSGYG 416
Query: 145 VHSRQFEVGVLDWFARLWELEKNEYWGYITNCGTEGNLHGILVGR 189
+S +FEV VLDWFA+LW++EK +YWGY+T GTEGNLHGIL+GR
Sbjct: 417 QNSVEFEVCVLDWFAKLWQMEKGDYWGYVTTGGTEGNLHGILMGR 551
>BU494238
Length = 448
Score = 123 bits (309), Expect = 6e-29
Identities = 58/105 (55%), Positives = 75/105 (71%), Gaps = 1/105 (0%)
Frame = +2
Query: 382 KCLRNAHYFK-DRLIEAGIGAMLNELSSTVVFERPHDEEFIRKWQLACKGNIAHVVVMPN 440
KC H D+L + IGAM NE S+TVVFERP D++F+RKW L+C+GN+AHVVVM +
Sbjct: 20 KCASQKHVTSLDKLHDNDIGAMRNEFSNTVVFERPPDDDFVRKWNLSCEGNVAHVVVMQH 199
Query: 441 VTIEKLDDFLNELVQKRATWFEDGTFQPYCIASDVGENSCLCAQH 485
VTIE LD F++ELV KR+ WF+ G +P CIA +VG +C C H
Sbjct: 200 VTIEMLDSFVSELVTKRSFWFQGGQRKPPCIADNVGGKNCACTLH 334
>BP071156
Length = 404
Score = 90.5 bits (223), Expect = 5e-19
Identities = 37/66 (56%), Positives = 51/66 (77%)
Frame = -3
Query: 420 FIRKWQLACKGNIAHVVVMPNVTIEKLDDFLNELVQKRATWFEDGTFQPYCIASDVGENS 479
F+RKW L+C+G++AHVVVM +VTIE +D F+ ELV+KR+ WF+ G +P CIA+ VG +
Sbjct: 402 FVRKWSLSCEGDVAHVVVMKHVTIEMIDSFVGELVKKRSIWFQGGQIKPPCIANTVGREN 223
Query: 480 CLCAQH 485
C CA H
Sbjct: 222 CACALH 205
>BP060635
Length = 360
Score = 64.3 bits (155), Expect = 4e-11
Identities = 29/53 (54%), Positives = 39/53 (72%)
Frame = -3
Query: 433 AHVVVMPNVTIEKLDDFLNELVQKRATWFEDGTFQPYCIASDVGENSCLCAQH 485
AHVVVM +VTIE +D F++ELV +R WF+DG P CIA+++G +C CA H
Sbjct: 358 AHVVVMQHVTIEMIDSFVSELVNQRTIWFQDGL--PPCIANNIGGENCACAVH 206
>TC14273 similar to UP|DCE1_ARATH (Q42521) Glutamate decarboxylase 1 (GAD
1) , partial (89%)
Length = 1826
Score = 60.5 bits (145), Expect = 6e-10
Identities = 71/269 (26%), Positives = 125/269 (46%), Gaps = 21/269 (7%)
Frame = +3
Query: 209 KAARMYRMECEKVETLNSGEIDCDDFKAKLLRHQDKPAIINVNIGTTVKGAVDDL----D 264
K AR + +E ++V+ L+ G D KA L ++ + + +G+T+ G +D+ D
Sbjct: 570 KFARYFEVELKEVK-LSEGYYVMDPQKAVDLVDENTICVAAI-LGSTLNGEFEDVKRLND 743
Query: 265 LVIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKRAPKVTFKKP-IGSVSVSGHKFVGCPM 323
L+I+K +E G+ IHVD A G + PF+ + F+ P + S++VSGHK+
Sbjct: 744 LLIEKNKETGWDTP---IHVDAASGGFIAPFIYPELEWDFRLPLVKSINVSGHKYGLVYA 914
Query: 324 PCGVQITRLEH--INALSRNVEYLASRDA--TIMGSRNGHAPIFLWYTLNRKGYRGFQKE 379
G I R + L ++ YL + T+ S+ I +Y L R GY G++
Sbjct: 915 GIGWVIWRNKEDLPEELIFHINYLGADQPTFTLNFSKGSSQIIAQYYQLIRLGYEGYKHV 1094
Query: 380 VQKCLRNAHYFKDRLIEAGIGAMLNE-----LSSTVVFERPHDEEFIRKWQLACKGNIAH 434
++ C N K+ L + G ++++ L + + + H EF L G I
Sbjct: 1095MENCRDNMLVLKEGLQKTGRFEIVSKDNGVPLVAFTLKDHTHYNEFQISDSLRRFGWIVP 1274
Query: 435 VVVMP----NVTIEKL---DDFLNELVQK 456
MP +VT+ ++ +DF L ++
Sbjct: 1275AYSMPPNTQHVTVVRVVIREDFSRTLAER 1361
>TC8050 similar to UP|Q8LPQ5 (Q8LPQ5) AT3g18290/MIE15_8, partial (5%)
Length = 506
Score = 29.3 bits (64), Expect = 1.5
Identities = 17/50 (34%), Positives = 24/50 (48%)
Frame = -2
Query: 154 VLDWFARLWELEKNEYWGYITNCGTEGNLHGILVGREVLPDGILYASRES 203
V DW R+W +N +WGY N G + G+ + G LY+S S
Sbjct: 199 VSDWITRVW*EPQNPHWGY--NQGKREVPFRLQSGQSMSGWGSLYSSGSS 56
>BP064478
Length = 509
Score = 28.1 bits (61), Expect = 3.4
Identities = 21/70 (30%), Positives = 36/70 (51%), Gaps = 2/70 (2%)
Frame = -1
Query: 125 QLQHFSINNLGDPFIESNYGVHSRQFEVGVLDWFARLWELEKNEYWGYITNCGTEGNLH- 183
Q+ H + + P ++ N+G ++R+ + L R L +N ++ Y+T GT +LH
Sbjct: 215 QVSHGTGLKVSIPQLQRNHGKNARKPFLLSLYQPPRTLSLSQNVFYSYLTR*GTFSSLHC 36
Query: 184 -GILVGREVL 192
ILV VL
Sbjct: 35 RSILVVNYVL 6
>TC7897 weakly similar to UP|ENL2_ARATH (Q9T076) Early nodulin-like protein
2 precursor (Phytocyanin-like protein), partial (31%)
Length = 1482
Score = 27.7 bits (60), Expect = 4.4
Identities = 29/82 (35%), Positives = 32/82 (38%), Gaps = 5/82 (6%)
Frame = -3
Query: 16 GAVELLPDDFDVSAIIKDPVP----PVVAADNGIGKEEAKINGGKEKREIVLGRNIHTTC 71
GAVELL DD VSA D VP AD G+ GG E G
Sbjct: 997 GAVELLDDDDGVSAGSDDGVPEGDTSGPEADGGVSPVVGDGAGGPPAGESPAGEG----- 833
Query: 72 LEV-TEPEADDEITGDRDAHMA 92
EV P A + + GD D A
Sbjct: 832 -EVAVGPPAGELLVGDEDGDSA 770
>TC13223
Length = 512
Score = 27.3 bits (59), Expect = 5.7
Identities = 11/24 (45%), Positives = 13/24 (53%)
Frame = -2
Query: 155 LDWFARLWELEKNEYWGYITNCGT 178
L WF R W + +YW I CGT
Sbjct: 112 LSWFIRTWLFQWPDYW--ICRCGT 47
>TC14164 weakly similar to UP|Q8VYH6 (Q8VYH6) AT4g17280/dl4675c, partial
(44%)
Length = 1606
Score = 26.6 bits (57), Expect = 9.8
Identities = 16/49 (32%), Positives = 26/49 (52%), Gaps = 4/49 (8%)
Frame = -1
Query: 27 VSAIIKDPVPPVV-AADNGIGKEEAKINGGKEKREIVLG---RNIHTTC 71
++A I P P+V D+G+ + K++GG KR + + R I T C
Sbjct: 373 ITARIDSPRDPLVRGGDSGVAESYVKLSGGLVKRPVKVRGKVRKITTRC 227
>TC11924 similar to UP|P93484 (P93484) BP-80 vacuolar sorting receptor,
partial (41%)
Length = 783
Score = 26.6 bits (57), Expect = 9.8
Identities = 16/59 (27%), Positives = 29/59 (49%), Gaps = 6/59 (10%)
Frame = +3
Query: 429 KGNIAHVVVMPNVTIE------KLDDFLNELVQKRATWFEDGTFQPYCIASDVGENSCL 481
KG+ V ++P + + KL+ +++ + FE+ T C++SDV N CL
Sbjct: 42 KGSRGDVTILPTLVVNNRQYRGKLEK--GAVMKAICSGFEETTEPAVCLSSDVETNECL 212
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.139 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,121,804
Number of Sequences: 28460
Number of extensions: 104704
Number of successful extensions: 452
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 449
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 450
length of query: 486
length of database: 4,897,600
effective HSP length: 94
effective length of query: 392
effective length of database: 2,222,360
effective search space: 871165120
effective search space used: 871165120
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Medicago: description of AC136472.15


