

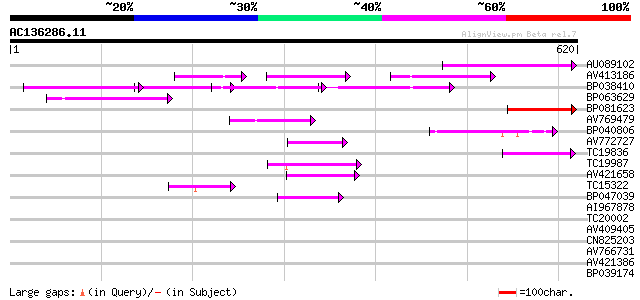
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136286.11 - phase: 0
(620 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AU089102 95 3e-20
AV413186 85 3e-17
BP038410 72 3e-13
BP063629 67 1e-11
BP081623 62 3e-10
AV769479 50 1e-06
BP040806 46 2e-05
AV772727 45 3e-05
TC19836 45 5e-05
TC19987 weakly similar to UP|Q7XJT3 (Q7XJT3) At2g17140 protein, ... 44 1e-04
AV421658 42 3e-04
TC15322 weakly similar to UP|Q9LG23 (Q9LG23) F14J16.14 (At1g5589... 42 4e-04
BP047039 41 7e-04
AI967878 40 0.001
TC20002 similar to UP|Q9FJY7 (Q9FJY7) Selenium-binding protein-l... 40 0.001
AV409405 39 0.002
CN825203 38 0.004
AV766731 36 0.016
AV421386 36 0.016
BP039174 35 0.047
>AU089102
Length = 480
Score = 95.1 bits (235), Expect = 3e-20
Identities = 48/148 (32%), Positives = 86/148 (57%), Gaps = 2/148 (1%)
Frame = +2
Query: 474 QPNSATFVSVLSACSHSGQIERGLRFF-RMIRKYGLDPKPEHFGCVVDLLGRAGQLGEAR 532
Q N T+V +L+ACSH+G ++ G+++F ++++ + K H+ C+VDL GRAG+L E
Sbjct: 32 QANDVTYVELLTACSHAGLVDEGIQYFDKLLKNRSIQVKEXHYACLVDLCGRAGRLKEXF 211
Query: 533 DLVQELA-EPPASVFDSLLGACRCYLDSNLGEEMAMKLIDIEPKNPAPLVVLSNIYAALG 591
+++ L + SV+ LL C + ++++G+ + K++ +E +N +LSN+YA+
Sbjct: 212 YIIEGLGVKLSLSVWGPLLAGCNVHGNADIGKLVTTKILKVEHENAGTYSLLSNMYASXR 391
Query: 592 RWSEVERIRGLITDKGLDKNSGISMIEV 619
W + + DKGL K IEV
Sbjct: 392 XWKXXAPVXMKMNDKGLKKQPRCXWIEV 475
>AV413186
Length = 398
Score = 85.1 bits (209), Expect = 3e-17
Identities = 47/116 (40%), Positives = 66/116 (56%), Gaps = 1/116 (0%)
Frame = +1
Query: 417 DTYMKCGCVSFARFVFDQFDVKPDDPAFWNAMIGGYGTNGDYESAFEVFYEMLDEMVQPN 476
D Y KCG + A VF+ K D WN MI +G A + +M+ V+PN
Sbjct: 4 DMYSKCGSLETAWRVFNLVGNK-QDVVLWNTMISALAHHGFGTEAMVMLNDMIRSGVEPN 180
Query: 477 SATFVSVLSACSHSGQIERGLRFFR-MIRKYGLDPKPEHFGCVVDLLGRAGQLGEA 531
ATFV +L+ACS SG ++ GL+FF+ M ++G+ P EH+ C+ DLL RAG E+
Sbjct: 181 RATFVVLLNACSLSGLVQEGLQFFKFMTSEFGVVPDQEHYACLTDLLVRAGYFDES 348
Score = 53.1 bits (126), Expect = 1e-07
Identities = 29/94 (30%), Positives = 52/94 (54%), Gaps = 2/94 (2%)
Frame = +1
Query: 281 DMYSKCGCWGSAFDVFSR-SEKRNLITWNSMIAGMMMNSESERAVELFERMVDEGILPDS 339
DMYSKCG +A+ VF+ K++++ WN+MI+ + + A+ + M+ G+ P+
Sbjct: 4 DMYSKCGSLETAWRVFNLVGNKQDVVLWNTMISALAHHGFGTEAMVMLNDMIRSGVEPNR 183
Query: 340 ATWNSLISGFAQKGVCVEAFKYFSKMQCA-GVAP 372
AT+ L++ + G+ E ++F M GV P
Sbjct: 184 ATFVVLLNACSLSGLVQEGLQFFKFMTSEFGVVP 285
Score = 42.0 bits (97), Expect = 3e-04
Identities = 27/80 (33%), Positives = 44/80 (54%), Gaps = 1/80 (1%)
Frame = +1
Query: 181 YSKCGVLVSSNKVFENLRVK-NVVTYNAFMSGLLQNGFHRVVFDVFKDMTMNLEEKPNKV 239
YSKCG L ++ +VF + K +VV +N +S L +GF + DM + E PN+
Sbjct: 10 YSKCGSLETAWRVFNLVGNKQDVVLWNTMISALAHHGFGTEAMVMLNDMIRSGVE-PNRA 186
Query: 240 TLVSVVSACATLSNIRLGKQ 259
T V +++AC+ ++ G Q
Sbjct: 187 TFVVLLNACSLSGLVQEGLQ 246
>BP038410
Length = 468
Score = 71.6 bits (174), Expect = 3e-13
Identities = 47/149 (31%), Positives = 70/149 (46%)
Frame = -2
Query: 338 DSATWNSLISGFAQKGVCVEAFKYFSKMQCAGVAPCLKILTSLLSVCGDSCVLRSAKAIH 397
D +WNS+ F F+KM +G+ + LL GD+ K++H
Sbjct: 404 DLVSWNSV-------------FDLFTKMCWSGLIASASTVLILLPAAGDTGNFVVGKSLH 264
Query: 398 GYALRICVDKDDFLATALVDTYMKCGCVSFARFVFDQFDVKPDDPAFWNAMIGGYGTNGD 457
GY ++I + TAL+D Y K G V + R +FD K D WN +IG Y NG
Sbjct: 263 GYYIQIGFSSHLNVLTALIDMYAKTGLVHWGRKIFDSLVDK--DVVLWNCLIGNYARNGL 90
Query: 458 YESAFEVFYEMLDEMVQPNSATFVSVLSA 486
A + +M + ++ NS+TFV +L A
Sbjct: 89 VGEALYLLQKMRIQGMKTNSSTFVGLLLA 3
Score = 60.8 bits (146), Expect = 6e-10
Identities = 37/131 (28%), Positives = 66/131 (50%)
Frame = -2
Query: 16 YKEALNLYSHLHSSSPTPNTFTFPILLKACSNLSSPSQTQILHAHLFKTGFHSHPHTSTA 75
+ +L++ + S + T ILL A + + + LH + + GF SH + TA
Sbjct: 392 WNSVFDLFTKMCWSGLIASASTVLILLPAAGDTGNFVVGKSLHGYYIQIGFSSHLNVLTA 213
Query: 76 LIASYAANTRSFHYALELFDEMPQPTITAFNAVLSGLSRNGPRGQAVWLFRQIGFWNIRP 135
LI YA T H+ ++FD + + +N ++ +RNG G+A++L +++ ++
Sbjct: 212 LIDMYA-KTGLVHWGRKIFDSLVDKDVVLWNCLIGNYARNGLVGEALYLLQKMRIQGMKT 36
Query: 136 NSVTIVSLLSA 146
NS T V LL A
Sbjct: 35 NSSTFVGLLLA 3
Score = 57.0 bits (136), Expect = 9e-09
Identities = 35/128 (27%), Positives = 67/128 (52%), Gaps = 2/128 (1%)
Frame = -2
Query: 221 VFDVFKDMTMNLEEKPNKVTLVSVVSACATLSNIRLGKQVHGLSMKLEACDHVMVVTSLV 280
VFD+F M + + T++ ++ A N +GK +HG +++ H+ V+T+L+
Sbjct: 383 VFDLFTKMCWS-GLIASASTVLILLPAAGDTGNFVVGKSLHGYYIQIGFSSHLNVLTALI 207
Query: 281 DMYSKCGC--WGSAFDVFSRSEKRNLITWNSMIAGMMMNSESERAVELFERMVDEGILPD 338
DMY+K G WG +F ++++ WN +I N A+ L ++M +G+ +
Sbjct: 206 DMYAKTGLVHWGR--KIFDSLVDKDVVLWNCLIGNYARNGLVGEALYLLQKMRIQGMKTN 33
Query: 339 SATWNSLI 346
S+T+ L+
Sbjct: 32 SSTFVGLL 9
Score = 40.4 bits (93), Expect = 9e-04
Identities = 28/111 (25%), Positives = 53/111 (47%)
Frame = -2
Query: 137 SVTIVSLLSARDVKNQSHVQQVHCLACKLGVEYDVYVSTSLVTAYSKCGVLVSSNKVFEN 196
S ++ L +A D N + +H ++G + V T+L+ Y+K G++ K+F++
Sbjct: 332 STVLILLPAAGDTGNFVVGKSLHGYYIQIGFSSHLNVLTALIDMYAKTGLVHWGRKIFDS 153
Query: 197 LRVKNVVTYNAFMSGLLQNGFHRVVFDVFKDMTMNLEEKPNKVTLVSVVSA 247
L K+VV +N + +NG + + M + K N T V ++ A
Sbjct: 152 LVDKDVVLWNCLIGNYARNGLVGEALYLLQKMRIQ-GMKTNSSTFVGLLLA 3
>BP063629
Length = 525
Score = 66.6 bits (161), Expect = 1e-11
Identities = 42/140 (30%), Positives = 74/140 (52%), Gaps = 2/140 (1%)
Frame = +1
Query: 41 LLKACSNLSSPSQTQILHAHLFKTGFHSHPHTSTALIA-SYAANTRSFHYALELFDEMPQ 99
LLK+C ++ Q Q L +F +G T L+A S ++ FHYAL +FD + Q
Sbjct: 109 LLKSCKSMCELKQIQAL---IFCSGLQQDRDTLNKLMAISTDSSIGDFHYALRIFDHIQQ 279
Query: 100 PTITAFNAVLSGLSRNGPRGQAVWLFRQIGFWNIRPNSVTIVSLLSARD-VKNQSHVQQV 158
P++ +N ++ ++ G +A+ LF+Q+ + P++ T +L A + + ++V
Sbjct: 280 PSLFNYNVMIKAFAKKGSFRRAISLFQQLREDGVWPDNYTYPYVLKAIGCLGDVGQGRKV 459
Query: 159 HCLACKLGVEYDVYVSTSLV 178
H K G+E+D YV SL+
Sbjct: 460 HAFVIKSGLEFDAYVCNSLM 519
Score = 40.0 bits (92), Expect = 0.001
Identities = 17/74 (22%), Positives = 36/74 (47%)
Frame = +1
Query: 4 DITVTKLVANGLYKEALNLYSHLHSSSPTPNTFTFPILLKACSNLSSPSQTQILHAHLFK 63
++ + G ++ A++L+ L P+ +T+P +LKA L Q + +HA + K
Sbjct: 298 NVMIKAFAKKGSFRRAISLFQQLREDGVWPDNYTYPYVLKAIGCLGDVGQGRKVHAFVIK 477
Query: 64 TGFHSHPHTSTALI 77
+G + +L+
Sbjct: 478 SGLEFDAYVCNSLM 519
>BP081623
Length = 461
Score = 62.0 bits (149), Expect = 3e-10
Identities = 28/75 (37%), Positives = 47/75 (62%)
Frame = -2
Query: 545 VFDSLLGACRCYLDSNLGEEMAMKLIDIEPKNPAPLVVLSNIYAALGRWSEVERIRGLIT 604
V+ +LLGACR Y D LG A L +++P++ ++L+N+YA + RW + ++R L+
Sbjct: 460 VWGALLGACRIYGDVGLGRWAAEHLFELKPQHCGYYILLANMYAEIERWDDANKVRQLMK 281
Query: 605 DKGLDKNSGISMIEV 619
+G KN G S ++V
Sbjct: 280 SRGAKKNPGCSWVKV 236
>AV769479
Length = 287
Score = 49.7 bits (117), Expect = 1e-06
Identities = 25/94 (26%), Positives = 50/94 (52%)
Frame = +1
Query: 241 LVSVVSACATLSNIRLGKQVHGLSMKLEACDHVMVVTSLVDMYSKCGCWGSAFDVFSRSE 300
LV ++ C+ ++ LGKQ+H +K + +++ + V+ Y++CG SAF F R
Sbjct: 4 LVCHMNLCSKRVDLALGKQIHAHILKSK-WRNLIADNAFVNFYAECGKISSAFRTFDRMA 180
Query: 301 KRNLITWNSMIAGMMMNSESERAVELFERMVDEG 334
KR+++ W ++I A+ + +M+ +G
Sbjct: 181 KRDVVCWTTIITACSQQGLGHEALLILSQMLGDG 282
Score = 28.5 bits (62), Expect = 3.3
Identities = 23/96 (23%), Positives = 40/96 (40%)
Frame = +1
Query: 376 ILTSLLSVCGDSCVLRSAKAIHGYALRICVDKDDFLATALVDTYMKCGCVSFARFVFDQF 435
+L +++C L K IH + L+ ++ A V+ Y +CG +S A FD+
Sbjct: 1 MLVCHMNLCSKRVDLALGKQIHAHILK-SKWRNLIADNAFVNFYAECGKISSAFRTFDRM 177
Query: 436 DVKPDDPAFWNAMIGGYGTNGDYESAFEVFYEMLDE 471
+ D W +I G A + +ML +
Sbjct: 178 AKR--DVVCWTTIITACSQQGLGHEALLILSQMLGD 279
>BP040806
Length = 534
Score = 46.2 bits (108), Expect = 2e-05
Identities = 41/151 (27%), Positives = 70/151 (46%), Gaps = 11/151 (7%)
Frame = +1
Query: 460 SAFEVFYEMLDE--MVQPNSATFVSVLSACSHSGQIERGLRFFRMIRKYGLDPKPEHFGC 517
S+ EVF +ML E +P T++ + SGQ R + F + + G DP PE +
Sbjct: 34 SSPEVF-DMLKEHSFYEPKEGTYMKLXVLLGKSGQPHRARQLFTTMIEEGCDPSPETYTA 210
Query: 518 VVDLLGRAGQLGEARDLVQE-----LAEPPASVFDSLLGAC----RCYLDSNLGEEMAMK 568
++ R+ L EA ++ E L +P + +L+ C + L L E+MA +
Sbjct: 211 LLAAYCRSNLLDEAFSILNEMKNHPLCQPDVFTYSTLIKPCVDAFKFDLVELLYEDMAAR 390
Query: 569 LIDIEPKNPAPLVVLSNIYAALGRWSEVERI 599
I P +VLS Y G++ ++E++
Sbjct: 391 --SIMPNTVTQNIVLSG-YGKAGKFDQMEKV 474
Score = 34.7 bits (78), Expect = 0.047
Identities = 18/72 (25%), Positives = 39/72 (54%), Gaps = 1/72 (1%)
Frame = +1
Query: 317 NSESERAVELFERMVDEGILPDSATWNSLISGFAQKGVCVEAFKYFSKMQCAGVA-PCLK 375
+ + RA +LF M++EG P T+ +L++ + + + EAF ++M+ + P +
Sbjct: 127 SGQPHRARQLFTTMIEEGCDPSPETYTALLAAYCRSNLLDEAFSILNEMKNHPLCQPDVF 306
Query: 376 ILTSLLSVCGDS 387
++L+ C D+
Sbjct: 307 TYSTLIKPCVDA 342
Score = 32.7 bits (73), Expect = 0.18
Identities = 24/104 (23%), Positives = 46/104 (44%), Gaps = 6/104 (5%)
Frame = +1
Query: 268 EACD-HVMVVTSLVDMYSKCGCWGSAFDVFSRSEKRNL-----ITWNSMIAGMMMNSESE 321
E CD T+L+ Y + AF + + + L T++++I + + +
Sbjct: 175 EGCDPSPETYTALLAAYCRSNLLDEAFSILNEMKNHPLCQPDVFTYSTLIKPCVDAFKFD 354
Query: 322 RAVELFERMVDEGILPDSATWNSLISGFAQKGVCVEAFKYFSKM 365
L+E M I+P++ T N ++SG+ + G + K S M
Sbjct: 355 LVELLYEDMAARSIMPNTVTQNIVLSGYGKAGKFDQMEKVLSSM 486
>AV772727
Length = 528
Score = 45.4 bits (106), Expect = 3e-05
Identities = 21/66 (31%), Positives = 38/66 (56%)
Frame = -2
Query: 304 LITWNSMIAGMMMNSESERAVELFERMVDEGILPDSATWNSLISGFAQKGVCVEAFKYFS 363
+IT++S++ + N ++A+ L ER+ D+GI PD T+N +I G + G +A + F
Sbjct: 527 VITYHSLLDALCKNHHVDKAIALLERVKDKGIQPDMYTYNIIIDGLCKSGRVKDAQELFQ 348
Query: 364 KMQCAG 369
+ G
Sbjct: 347 DLLIKG 330
Score = 36.6 bits (83), Expect = 0.012
Identities = 31/105 (29%), Positives = 51/105 (48%), Gaps = 20/105 (19%)
Frame = -2
Query: 138 VTIVSLLSARDVKNQSHVQQVHCLACKL---GVEYDVYVSTSLVTAYSKCGVLVSSNKVF 194
+T SLL A KN HV + L ++ G++ D+Y ++ K G + + ++F
Sbjct: 524 ITYHSLLDAL-CKNH-HVDKAIALLERVKDKGIQPDMYTYNIIIDGLCKSGRVKDAQELF 351
Query: 195 ENLRVK----NVVTYNAFMSG-------------LLQNGFHRVVF 222
++L +K NVVTYN ++G L+QNG H + F
Sbjct: 350 QDLLIKGYRLNVVTYNIMINGLCIEGLSDEALALLIQNGRHALCF 216
Score = 32.0 bits (71), Expect = 0.30
Identities = 16/56 (28%), Positives = 33/56 (58%)
Frame = -2
Query: 303 NLITWNSMIAGMMMNSESERAVELFERMVDEGILPDSATWNSLISGFAQKGVCVEA 358
++ T+N +I G+ + + A ELF+ ++ +G + T+N +I+G +G+ EA
Sbjct: 425 DMYTYNIIIDGLCKSGRVKDAQELFQDLLIKGYRLNVVTYNIMINGLCIEGLSDEA 258
>TC19836
Length = 464
Score = 44.7 bits (104), Expect = 5e-05
Identities = 25/79 (31%), Positives = 41/79 (51%)
Frame = +2
Query: 540 EPPASVFDSLLGACRCYLDSNLGEEMAMKLIDIEPKNPAPLVVLSNIYAALGRWSEVERI 599
+P ++ SL+ AC+ + ++ E + +LI EP N A +L+ IYA G W E++
Sbjct: 17 KPSTRIWSSLVSACKLHGRLDIAEMLGPQLIRSEPDNAANYTLLNMIYAEHGHWLHKEQV 196
Query: 600 RGLITDKGLDKNSGISMIE 618
+ + L K G S IE
Sbjct: 197 MEAMKLQRLKKCYGFSRIE 253
>TC19987 weakly similar to UP|Q7XJT3 (Q7XJT3) At2g17140 protein, partial
(12%)
Length = 350
Score = 43.5 bits (101), Expect = 1e-04
Identities = 29/106 (27%), Positives = 48/106 (44%), Gaps = 4/106 (3%)
Frame = +3
Query: 283 YSKCGCWGSAFDVFSRSEK----RNLITWNSMIAGMMMNSESERAVELFERMVDEGILPD 338
+ K G SA V E+ + L T+NS+I G+ + L + M + GI PD
Sbjct: 30 FCKEGKISSALRVLKDMERNGCSKTLQTYNSLILGLGSKGQIFEMYGLMDEMRERGICPD 209
Query: 339 SATWNSLISGFAQKGVCVEAFKYFSKMQCAGVAPCLKILTSLLSVC 384
T+N++IS + G +A +M G++P + L+ C
Sbjct: 210 ICTYNNVISCLCEGGKTEDATSLLHEMLDKGISPNISSFKILIKSC 347
Score = 32.0 bits (71), Expect = 0.30
Identities = 18/78 (23%), Positives = 35/78 (44%)
Frame = +3
Query: 305 ITWNSMIAGMMMNSESERAVELFERMVDEGILPDSATWNSLISGFAQKGVCVEAFKYFSK 364
+T+++ I + A+ + + M G T+NSLI G KG E + +
Sbjct: 3 VTYDTFIWKFCKEGKISSALRVLKDMERNGCSKTLQTYNSLILGLGSKGQIFEMYGLMDE 182
Query: 365 MQCAGVAPCLKILTSLLS 382
M+ G+ P + +++S
Sbjct: 183 MRERGICPDICTYNNVIS 236
Score = 30.0 bits (66), Expect = 1.1
Identities = 14/47 (29%), Positives = 25/47 (52%)
Frame = +3
Query: 441 DPAFWNAMIGGYGTNGDYESAFEVFYEMLDEMVQPNSATFVSVLSAC 487
D +N +I G E A + +EMLD+ + PN ++F ++ +C
Sbjct: 207 DICTYNNVISCLCEGGKTEDATSLLHEMLDKGISPNISSFKILIKSC 347
>AV421658
Length = 281
Score = 42.0 bits (97), Expect = 3e-04
Identities = 25/82 (30%), Positives = 46/82 (55%), Gaps = 2/82 (2%)
Frame = +1
Query: 303 NLITWNSMIAGM--MMNSESERAVELFERMVDEGILPDSATWNSLISGFAQKGVCVEAFK 360
+++T+N++I+ + + E+A E+ MV +GILPD+ T+ LI + EA+
Sbjct: 31 DVVTYNTLISKFCKLKEPDLEKAFEMKAEMVHKGILPDADTYEPLIRTLCLQQRLSEAYD 210
Query: 361 YFSKMQCAGVAPCLKILTSLLS 382
F +M GV+P + T L++
Sbjct: 211 LFREMLRWGVSPNNETYTGLMN 276
>TC15322 weakly similar to UP|Q9LG23 (Q9LG23) F14J16.14
(At1g55890/F14J16_4), partial (33%)
Length = 588
Score = 41.6 bits (96), Expect = 4e-04
Identities = 22/78 (28%), Positives = 45/78 (57%), Gaps = 4/78 (5%)
Frame = +2
Query: 174 STSLVTAYSKCGVLVSSNKVFENLRVKN----VVTYNAFMSGLLQNGFHRVVFDVFKDMT 229
S L+T Y K G+ + ++F+ + +N V+++NA ++ L + +RVV +FK++
Sbjct: 350 SARLITLYGKSGMPKRARQMFDEMPERNCTRTVLSFNALLAAYLHSKQYRVVERIFKEVP 529
Query: 230 MNLEEKPNKVTLVSVVSA 247
+ KP+ V+ +V+ A
Sbjct: 530 GEISVKPDLVSYNTVIKA 583
>BP047039
Length = 527
Score = 40.8 bits (94), Expect = 7e-04
Identities = 22/72 (30%), Positives = 37/72 (50%)
Frame = -3
Query: 294 DVFSRSEKRNLITWNSMIAGMMMNSESERAVELFERMVDEGILPDSATWNSLISGFAQKG 353
D+F + N T+N+MI G+ + A+ L +M D G + D T+ ++IS +KG
Sbjct: 525 DLFKKGYNLNNWTYNTMINGLCKEGLFDEALTLMSKMEDNGCIHDPITFETIISALFEKG 346
Query: 354 VCVEAFKYFSKM 365
+A K +M
Sbjct: 345 ENDKAEKLLREM 310
>AI967878
Length = 393
Score = 39.7 bits (91), Expect = 0.001
Identities = 27/96 (28%), Positives = 43/96 (44%), Gaps = 21/96 (21%)
Frame = +1
Query: 302 RNLITWNSMIAGMMMNSESERAVELFERMV-----------------DEGIL---PDSAT 341
RNL++WN+MI G ++ E + LF+ MV D G++ PD T
Sbjct: 31 RNLVSWNAMILGHCIHGNPEDGLSLFDEMVGMDKVKGEVEIDESPCADRGVVRLPPDEVT 210
Query: 342 WNSLISGFAQKGVCVEAFKYFSKM-QCAGVAPCLKI 376
+ ++ A+ + E YF +M G+ P L I
Sbjct: 211 FIGILCACARAELLAEGRSYFKQMTDVFGLKPILLI 318
Score = 37.4 bits (85), Expect = 0.007
Identities = 25/87 (28%), Positives = 40/87 (45%), Gaps = 21/87 (24%)
Frame = +1
Query: 445 WNAMIGGYGTNGDYESAFEVFYEM-----------LDE---------MVQPNSATFVSVL 484
WNAMI G+ +G+ E +F EM +DE + P+ TF+ +L
Sbjct: 46 WNAMILGHCIHGNPEDGLSLFDEMVGMDKVKGEVEIDESPCADRGVVRLPPDEVTFIGIL 225
Query: 485 SACSHSGQIERGLRFFR-MIRKYGLDP 510
AC+ + + G +F+ M +GL P
Sbjct: 226 CACARAELLAEGRSYFKQMTDVFGLKP 306
>TC20002 similar to UP|Q9FJY7 (Q9FJY7) Selenium-binding protein-like,
partial (4%)
Length = 560
Score = 39.7 bits (91), Expect = 0.001
Identities = 27/112 (24%), Positives = 53/112 (47%), Gaps = 4/112 (3%)
Frame = -3
Query: 37 TFPILLKACSNLSSPSQTQILHAHLFKTGFHSHPHTSTALIASYAANTRS--FHYALELF 94
T L CS ++ + +HA +++TGFH + +I A + + +YA+ +F
Sbjct: 348 TLMTLFNRCSTMNHLKE---IHARIYQTGFHQNHLVVGKIIVFCAVSVPAGDMNYAVSVF 178
Query: 95 DEMPQPTITAFNAVLSGLSRNGPRGQAVWLFR--QIGFWNIRPNSVTIVSLL 144
D + +P +N ++ G +AV ++ Q G ++ P++ T LL
Sbjct: 177 DRVDKPDAFLWNTMIRGFGNTNQPEKAVLFYKRMQQGEPHVVPDTFTFSFLL 22
Score = 33.5 bits (75), Expect = 0.10
Identities = 26/66 (39%), Positives = 38/66 (57%), Gaps = 4/66 (6%)
Frame = -3
Query: 423 GCVSFARFVFDQFDVKPDDPAF-WNAMIGGYGTNGDYESAFEVFYEMLDE---MVQPNSA 478
G +++A VFD+ D KPD AF WN MI G+G E A +FY+ + + V P++
Sbjct: 207 GDMNYAVSVFDRVD-KPD--AFLWNTMIRGFGNTNQPEKAV-LFYKRMQQGEPHVVPDTF 40
Query: 479 TFVSVL 484
TF +L
Sbjct: 39 TFSFLL 22
Score = 29.6 bits (65), Expect = 1.5
Identities = 16/44 (36%), Positives = 26/44 (58%)
Frame = -3
Query: 323 AVELFERMVDEGILPDSATWNSLISGFAQKGVCVEAFKYFSKMQ 366
AV +F+R VD+ PD+ WN++I GF +A ++ +MQ
Sbjct: 192 AVSVFDR-VDK---PDAFLWNTMIRGFGNTNQPEKAVLFYKRMQ 73
>AV409405
Length = 424
Score = 38.9 bits (89), Expect = 0.002
Identities = 28/88 (31%), Positives = 41/88 (45%)
Frame = +1
Query: 41 LLKACSNLSSPSQTQILHAHLFKTGFHSHPHTSTALIASYAANTRSFHYALELFDEMPQP 100
LL+ C S SQ LH+ K G T L Y + S H+A +LFDE P
Sbjct: 163 LLETCHCEKSTSQ---LHSLCLKLGLTHDSFIVTKLNVLYGKHA-SIHHAHKLFDETPDK 330
Query: 101 TITAFNAVLSGLSRNGPRGQAVWLFRQI 128
++ +NA+L G + + LFR++
Sbjct: 331 SVYLWNALLRRYRAEGEWAETLSLFRRM 414
Score = 38.1 bits (87), Expect = 0.004
Identities = 20/77 (25%), Positives = 36/77 (45%)
Frame = +1
Query: 152 QSHVQQVHCLACKLGVEYDVYVSTSLVTAYSKCGVLVSSNKVFENLRVKNVVTYNAFMSG 211
+ Q+H L KLG+ +D ++ T L Y K + ++K+F+ K+V +NA +
Sbjct: 184 EKSTSQLHSLCLKLGLTHDSFIVTKLNVLYGKHASIHHAHKLFDETPDKSVYLWNALLRR 363
Query: 212 LLQNGFHRVVFDVFKDM 228
G +F+ M
Sbjct: 364 YRAEGEWAETLSLFRRM 414
>CN825203
Length = 644
Score = 38.1 bits (87), Expect = 0.004
Identities = 28/122 (22%), Positives = 58/122 (46%), Gaps = 6/122 (4%)
Frame = -2
Query: 308 NSMIAGMMMNSESERAVELFERMVDEGILPDSATWNSLISGFAQKGVCVEAFKYFSK--- 364
N ++ G E+A + MVD+G P +W+ + SG K +AF+ F +
Sbjct: 634 NILLIGYSRKGLIEKAETMLRSMVDKGKTPTPNSWSIIASGHVAKENMEKAFQCFKEALA 455
Query: 365 --MQCAGVAPCLKILTSLLSVCGDSCVLRSAK-AIHGYALRICVDKDDFLATALVDTYMK 421
+ G P +++S+LS D+ + + ++ + +++D +L +L+ Y++
Sbjct: 454 VLAENKGWRPKSDVVSSILSWVSDNRDIEEVEDFVNSLKKVMSMNRDMYL--SLIKLYVR 281
Query: 422 CG 423
CG
Sbjct: 280 CG 275
>AV766731
Length = 576
Score = 36.2 bits (82), Expect = 0.016
Identities = 43/201 (21%), Positives = 73/201 (35%), Gaps = 2/201 (0%)
Frame = -3
Query: 303 NLITWNSMIAGMMMNSESERAVELFERMVDEGILPDSATWNSLISGFAQKGVCVEAFKYF 362
N + NS+I G+ AVEL + M + G D T NSL G + + +A F
Sbjct: 550 NTVKKNSLINGLCKARRISCAVELVDVMHETGPPADLITTNSLFDGLCKHHLLDKAIALF 371
Query: 363 SKMQCAGVAPCLKILTSLLSVCGDSCVLRSAKAIHGYALRICVDKDDFLATALVDTYMKC 422
K++ + P I Y + ++D K
Sbjct: 370 MKVKDPKIQP----------------------NIPPYPV-------------IIDGLCKV 296
Query: 423 GCVSFARFVFDQF--DVKPDDPAFWNAMIGGYGTNGDYESAFEVFYEMLDEMVQPNSATF 480
G + A+ +F + + + MI GY G + A + +M D PN+ F
Sbjct: 295 GRLKIAQEIFQVLLSEGYNLNVMTYTVMINGYCKEGLLDEAQALLSKMEDNGCIPNAVNF 116
Query: 481 VSVLSACSHSGQIERGLRFFR 501
+++ A + ++ R FR
Sbjct: 115 QTIICALFEKNENDKAERLFR 53
Score = 34.3 bits (77), Expect = 0.061
Identities = 30/141 (21%), Positives = 61/141 (42%), Gaps = 4/141 (2%)
Frame = -3
Query: 236 PNKVTLVSVVSACATLSNIRLGKQVHGLSMKLEACDHVMVVTSLVDMYSKCGCWGSAFDV 295
PN V S+++ I ++ + + ++ SL D K A +
Sbjct: 553 PNTVKKNSLINGLCKARRISCAVELVDVMHETGPPADLITTNSLFDGLCKHHLLDKAIAL 374
Query: 296 FSRSE----KRNLITWNSMIAGMMMNSESERAVELFERMVDEGILPDSATWNSLISGFAQ 351
F + + + N+ + +I G+ + A E+F+ ++ EG + T+ +I+G+ +
Sbjct: 373 FMKVKDPKIQPNIPPYPVIIDGLCKVGRLKIAQEIFQVLLSEGYNLNVMTYTVMINGYCK 194
Query: 352 KGVCVEAFKYFSKMQCAGVAP 372
+G+ EA SKM+ G P
Sbjct: 193 EGLLDEAQALLSKMEDNGCIP 131
>AV421386
Length = 312
Score = 36.2 bits (82), Expect = 0.016
Identities = 17/52 (32%), Positives = 30/52 (57%)
Frame = +1
Query: 459 ESAFEVFYEMLDEMVQPNSATFVSVLSACSHSGQIERGLRFFRMIRKYGLDP 510
+ A + F EMLD +PNSA + +++ +G+I+ FF+ + K G+ P
Sbjct: 145 DEALKFFEEMLDYQCKPNSAIYNILINGFGKAGKIDIACDFFKRMVKEGIRP 300
Score = 35.4 bits (80), Expect = 0.027
Identities = 17/95 (17%), Positives = 41/95 (42%)
Frame = +1
Query: 446 NAMIGGYGTNGDYESAFEVFYEMLDEMVQPNSATFVSVLSACSHSGQIERGLRFFRMIRK 505
N +I + A +++YE++ P T+ + + + + L+FF +
Sbjct: 1 NIIISALVKSNSLNKALDLYYELISGDFSPTPCTYGPFIDGPLKAERCDEALKFFEEMLD 180
Query: 506 YGLDPKPEHFGCVVDLLGRAGQLGEARDLVQELAE 540
Y P + +++ G+AG++ A D + + +
Sbjct: 181 YQCKPNSAIYNILINGFGKAGKIDIACDFFKRMVK 285
Score = 29.3 bits (64), Expect = 2.0
Identities = 15/38 (39%), Positives = 21/38 (54%)
Frame = +1
Query: 301 KRNLITWNSMIAGMMMNSESERAVELFERMVDEGILPD 338
K N +N +I G + + A + F+RMV EGI PD
Sbjct: 190 KPNSAIYNILINGFGKAGKIDIACDFFKRMVKEGIRPD 303
>BP039174
Length = 564
Score = 34.7 bits (78), Expect = 0.047
Identities = 28/98 (28%), Positives = 39/98 (39%), Gaps = 3/98 (3%)
Frame = -2
Query: 414 ALVDTYMKCGCVSFARFVFDQFDVK---PDDPAFWNAMIGGYGTNGDYESAFEVFYEMLD 470
AL+ Y + G A+ VFD + PDD +I YG G E A F M
Sbjct: 545 ALLRAYSRIGNAEGAQRVFDAIQLAGIIPDDK-ICGLVIKAYGMAGQSEKARIAFENMKR 369
Query: 471 EMVQPNSATFVSVLSACSHSGQIERGLRFFRMIRKYGL 508
++P SVL A ++ L F + K G+
Sbjct: 368 AGIEPTDRCIGSVLVAYEKESKLNTALEFLIDLEKEGI 255
Score = 29.6 bits (65), Expect = 1.5
Identities = 17/62 (27%), Positives = 30/62 (47%)
Frame = -2
Query: 320 SERAVELFERMVDEGILPDSATWNSLISGFAQKGVCVEAFKYFSKMQCAGVAPCLKILTS 379
+E A +F+ + GI+PD +I + G +A F M+ AG+ P + + S
Sbjct: 512 AEGAQRVFDAIQLAGIIPDDKICGLVIKAYGMAGQSEKARIAFENMKRAGIEPTDRCIGS 333
Query: 380 LL 381
+L
Sbjct: 332 VL 327
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.322 0.135 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,071,572
Number of Sequences: 28460
Number of extensions: 165993
Number of successful extensions: 1056
Number of sequences better than 10.0: 66
Number of HSP's better than 10.0 without gapping: 991
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1043
length of query: 620
length of database: 4,897,600
effective HSP length: 96
effective length of query: 524
effective length of database: 2,165,440
effective search space: 1134690560
effective search space used: 1134690560
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC136286.11


