

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136140.11 - phase: 0 /pseudo
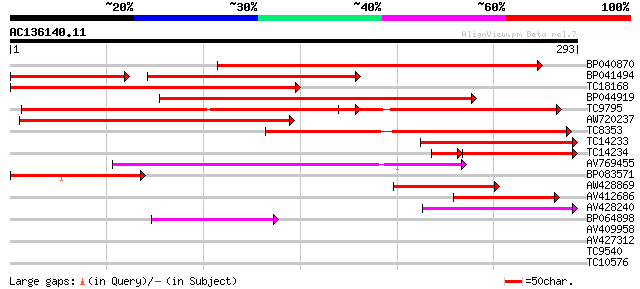
(293 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BP040870 315 4e-87
BP041494 191 3e-78
TC18168 homologue to UP|Q8GSN4 (Q8GSN4) Non-cell-autonomous heat... 285 8e-78
BP044919 249 5e-67
TC9795 homologue to UP|Q9ATB8 (Q9ATB8) BiP-isoform D (Fragment),... 236 3e-63
AW720237 221 1e-58
TC8353 homologue to UP|HS7M_PHAVU (Q01899) Heat shock 70 kDa pro... 189 5e-49
TC14233 homologue to UP|Q41027 (Q41027) PsHSC71.0, partial (67%) 161 1e-40
TC14234 homologue to UP|Q41027 (Q41027) PsHSC71.0, partial (66%) 120 8e-33
AV769455 127 2e-30
BP083571 115 1e-26
AW428869 79 9e-16
AV412686 74 2e-14
AV428240 61 3e-10
BP064898 53 7e-08
AV409958 35 0.019
AV427312 30 0.48
TC9540 26 9.1
TC10576 similar to UP|Q93ZQ3 (Q93ZQ3) AT3g63200/F16M2_50, partia... 26 9.1
>BP040870
Length = 505
Score = 315 bits (808), Expect = 4e-87
Identities = 157/168 (93%), Positives = 166/168 (98%)
Frame = +2
Query: 108 GVNYKGEDKQFAAEEISSMVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGV 167
GVNYKGE+KQF+AEEISSMVL+KM+EIAEAYLG+T+KNAVVTVPAYFNDSQRQATKDAGV
Sbjct: 2 GVNYKGEEKQFSAEEISSMVLMKMKEIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGV 181
Query: 168 IAGLNVLRIINEPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKA 227
I+GLNV+RIINEPTAAAIAYGLDKKATS GEKNVLIFDLGGGTFDVSLLTIEEGIFEVK+
Sbjct: 182 ISGLNVMRIINEPTAAAIAYGLDKKATSTGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKS 361
Query: 228 TAGDTHLGGEDFDNRMVNHFVQEFKRKNKKDISGNPRALRRLRTACER 275
TAGDTHLGGEDFDNRMVNHFVQEFKRKNKKDISGN RALRRLRTACER
Sbjct: 362 TAGDTHLGGEDFDNRMVNHFVQEFKRKNKKDISGNARALRRLRTACER 505
>BP041494
Length = 572
Score = 191 bits (486), Expect(2) = 3e-78
Identities = 96/110 (87%), Positives = 103/110 (93%)
Frame = +3
Query: 72 DAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAAEEISSMVLIKM 131
DAKRLIGR+ SD SV+SDMKLWPFKVI+GP ++PMI VNYK E+KQF AEEISSMVLIKM
Sbjct: 243 DAKRLIGRKFSDDSVESDMKLWPFKVISGPVDRPMIVVNYKNEEKQFTAEEISSMVLIKM 422
Query: 132 REIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPT 181
REIAEAYLGSTVKN VVTVPAYFNDSQRQATKDAGVIAGLNV+RIINEPT
Sbjct: 423 REIAEAYLGSTVKNVVVTVPAYFNDSQRQATKDAGVIAGLNVMRIINEPT 572
Score = 117 bits (292), Expect(2) = 3e-78
Identities = 55/62 (88%), Positives = 58/62 (92%)
Frame = +1
Query: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
M G+GE PAIGIDLGTTYSCVGVWQ+DRVEII NDQGNRTTPSYVAFTD ERLIGDAAKN
Sbjct: 61 MVGEGESPAIGIDLGTTYSCVGVWQNDRVEIIPNDQGNRTTPSYVAFTDFERLIGDAAKN 240
Query: 61 QV 62
Q+
Sbjct: 241 QM 246
>TC18168 homologue to UP|Q8GSN4 (Q8GSN4) Non-cell-autonomous heat shock
cognate protein 70, partial (23%)
Length = 508
Score = 285 bits (728), Expect = 8e-78
Identities = 140/150 (93%), Positives = 147/150 (97%)
Frame = +2
Query: 1 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKN 60
MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTD+ERLIGDAAKN
Sbjct: 59 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAKN 238
Query: 61 QVAMNPVNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAA 120
QVAMNP+NTVFDAKRLIGRR+SDASVQSDMKLWPFKV +GP EKPMI V+YKGEDKQFAA
Sbjct: 239 QVAMNPINTVFDAKRLIGRRVSDASVQSDMKLWPFKVNSGPAEKPMIQVSYKGEDKQFAA 418
Query: 121 EEISSMVLIKMREIAEAYLGSTVKNAVVTV 150
EEISSMVL+KMREIAEAYLGST+KNAVVTV
Sbjct: 419 EEISSMVLMKMREIAEAYLGSTIKNAVVTV 508
>BP044919
Length = 519
Score = 249 bits (635), Expect = 5e-67
Identities = 123/164 (75%), Positives = 144/164 (87%)
Frame = +3
Query: 78 GRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAAEEISSMVLIKMREIAEA 137
GR+ SD+ +Q D+KLWPFKVIAG +KPMI V +KGE+K+ EEISSMVL KM+EIAEA
Sbjct: 27 GRKYSDSVIQEDVKLWPFKVIAGTDDKPMIVVKHKGEEKKLVPEEISSMVLSKMKEIAEA 206
Query: 138 YLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAIAYGLDKKATSVG 197
+L S+VKNAVVTVPAYFND+QR+ATKDAG IAGLNV+RIINEPTAAA+AYGL K+A VG
Sbjct: 207 FLESSVKNAVVTVPAYFNDAQRKATKDAGTIAGLNVMRIINEPTAAALAYGLQKRANLVG 386
Query: 198 EKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDN 241
E+NV IFDLGGGTFDVSLLTI++ FEVKAT+GDTHLG +DFDN
Sbjct: 387 ERNVFIFDLGGGTFDVSLLTIKDDNFEVKATSGDTHLGXKDFDN 518
>TC9795 homologue to UP|Q9ATB8 (Q9ATB8) BiP-isoform D (Fragment), partial
(63%)
Length = 981
Score = 236 bits (602), Expect = 3e-63
Identities = 121/176 (68%), Positives = 141/176 (79%), Gaps = 1/176 (0%)
Frame = +1
Query: 7 GPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNP 66
G IGIDLGTTYSCVGV+++ VEIIANDQGNR TPS+VAF D ERLIG+AAKNQ A+NP
Sbjct: 145 GTVIGIDLGTTYSCVGVYRNGHVEIIANDQGNRITPSWVAFNDGERLIGEAAKNQFAVNP 324
Query: 67 VNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYK-GEDKQFAAEEISS 125
T+FD KRLIGR+ D VQ DMKL P+K++ G KP I V K GE K F+ EE+S+
Sbjct: 325 ERTIFDVKRLIGRKFEDKEVQRDMKLVPYKIVNKDG-KPYIQVKIKDGETKVFSPEEVSA 501
Query: 126 MVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPT 181
M+L KM+E AEA+LG + +AVVTVPAYFND+QRQATKDAGVIAGLNV RIINEPT
Sbjct: 502 MILTKMKETAEAFLGKKINDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPT 669
Score = 140 bits (352), Expect = 3e-34
Identities = 72/115 (62%), Positives = 88/115 (75%)
Frame = +2
Query: 171 LNVLRIINEPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAG 230
L +L + P AAAIAYGLDKK GEKN+L+FDLGGGTFDVS+LTI+ G+FEV AT G
Sbjct: 638 LMLLELSMNPPAAAIAYGLDKKG---GEKNILVFDLGGGTFDVSILTIDNGVFEVLATNG 808
Query: 231 DTHLGGEDFDNRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQ 285
DTHLGGEDFD R++ +F++ K+K+ KDIS + RAL +LR ERAKR LSS Q
Sbjct: 809 DTHLGGEDFDQRIMEYFIKLIKKKHGKDISKDNRALGKLRREAERAKRALSSQHQ 973
>AW720237
Length = 459
Score = 221 bits (562), Expect = 1e-58
Identities = 107/142 (75%), Positives = 120/142 (84%)
Frame = +3
Query: 6 EGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMN 65
+ AIGIDLGTTYSCVGVW +DRVEIIANDQGNRTTPSYV FT+SERLIGDAAKNQVA N
Sbjct: 33 QATAIGIDLGTTYSCVGVWMNDRVEIIANDQGNRTTPSYVGFTESERLIGDAAKNQVARN 212
Query: 66 PVNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAAEEISS 125
P+NTVFDAKRLIGR+ D VQ D+KLWPFKV AGP KP+I V YKGE K+F AE++SS
Sbjct: 213 PINTVFDAKRLIGRKFDDPIVQKDIKLWPFKVEAGPDGKPLIAVKYKGETKKFHAEQVSS 392
Query: 126 MVLIKMREIAEAYLGSTVKNAV 147
MVL KM+E AEAYL T+K+ V
Sbjct: 393 MVLTKMKETAEAYLNKTIKDIV 458
>TC8353 homologue to UP|HS7M_PHAVU (Q01899) Heat shock 70 kDa protein,
mitochondrial precursor, partial (74%)
Length = 1752
Score = 189 bits (480), Expect = 5e-49
Identities = 99/158 (62%), Positives = 119/158 (74%)
Frame = +3
Query: 133 EIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAIAYGLDKK 192
E AEAYLG +V AV+TVPAYFND+ RQATKDAG IAGL+V RIINEPTAAA++YG+ K
Sbjct: 3 ETAEAYLGKSVSKAVITVPAYFNDAHRQATKDAGRIAGLDVQRIINEPTAAALSYGMTNK 182
Query: 193 ATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFVQEFK 252
E + +FDLGGGTFDVS+L I G+FEVKAT GDT LGGEDFDN +++ V EFK
Sbjct: 183 -----EGLIAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLVDEFK 347
Query: 253 RKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEI 290
R D+S + AL+RLR A E+AK LSST+QT I +
Sbjct: 348 RTESIDLSKDKLALQRLREAAEKAKIELSSTSQTEINL 461
>TC14233 homologue to UP|Q41027 (Q41027) PsHSC71.0, partial (67%)
Length = 1570
Score = 161 bits (408), Expect = 1e-40
Identities = 81/81 (100%), Positives = 81/81 (100%)
Frame = +1
Query: 213 VSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFVQEFKRKNKKDISGNPRALRRLRTA 272
VSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFVQEFKRKNKKDISGNPRALRRLRTA
Sbjct: 1 VSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFVQEFKRKNKKDISGNPRALRRLRTA 180
Query: 273 CERAKRTLSSTAQTTIEIDSL 293
CERAKRTLSSTAQTTIEIDSL
Sbjct: 181 CERAKRTLSSTAQTTIEIDSL 243
>TC14234 homologue to UP|Q41027 (Q41027) PsHSC71.0, partial (66%)
Length = 1528
Score = 120 bits (301), Expect(2) = 8e-33
Identities = 59/59 (100%), Positives = 59/59 (100%)
Frame = +3
Query: 235 GGEDFDNRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSL 293
GGEDFDNRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSL
Sbjct: 48 GGEDFDNRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSL 224
Score = 36.2 bits (82), Expect(2) = 8e-33
Identities = 16/16 (100%), Positives = 16/16 (100%)
Frame = +1
Query: 219 EEGIFEVKATAGDTHL 234
EEGIFEVKATAGDTHL
Sbjct: 1 EEGIFEVKATAGDTHL 48
>AV769455
Length = 556
Score = 127 bits (319), Expect = 2e-30
Identities = 71/186 (38%), Positives = 105/186 (56%), Gaps = 3/186 (1%)
Frame = +1
Query: 54 IGDAAKNQVAMNPVNTVFDAKRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKG 113
+G A + MNP N++ KRLIGR+ +D +Q D+K PF V GP P+I Y G
Sbjct: 4 LGTAGASSTMMNPKNSISQLKRLIGRQFADPELQRDLKSLPFAVTEGPDGFPLIHARYLG 183
Query: 114 EDKQFAAEEISSMVLIKMREIAEAYLGSTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNV 173
E + F ++ M+L ++EIA+ L + V + + +P YF D QR+A DA IAGL+
Sbjct: 184 ETRTFTPTQVFGMMLSNLKEIAQKNLNAAVVDCCIGIPIYFTDLQRRAVLDAATIAGLHP 363
Query: 174 LRIINEPTAAAIAYGLDKKATSVGEK---NVLIFDLGGGTFDVSLLTIEEGIFEVKATAG 230
LR+ +E TA A+AYG+ K T + E NV D+G + V + ++G +V A +
Sbjct: 364 LRLFHETTATALAYGIYK--TDLPENDPLNVAFVDVGHASMQVCIAGYKKGQLKVLAHSY 537
Query: 231 DTHLGG 236
D LGG
Sbjct: 538 DRSLGG 555
>BP083571
Length = 423
Score = 115 bits (287), Expect = 1e-26
Identities = 57/72 (79%), Positives = 62/72 (85%), Gaps = 2/72 (2%)
Frame = +1
Query: 1 MAGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 58
MA K EGPAIGIDLGTTYSCV VWQ +DR EII N+QGNRTTPS+VAFTD++RLIGDAA
Sbjct: 145 MATKYEGPAIGIDLGTTYSCVAVWQEQNDRAEIIHNEQGNRTTPSFVAFTDTQRLIGDAA 324
Query: 59 KNQVAMNPVNTV 70
KNQ A NP NTV
Sbjct: 325 KNQAATNPTNTV 360
>AW428869
Length = 181
Score = 79.0 bits (193), Expect = 9e-16
Identities = 33/55 (60%), Positives = 46/55 (83%)
Frame = +1
Query: 199 KNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFVQEFKR 253
+ +L+FDLGGGTFDVS+L + +G+FEV +T+GDTHLGG+DFD R+V+ +FKR
Sbjct: 16 ETILVFDLGGGTFDVSVLEVGDGVFEVLSTSGDTHLGGDDFDKRIVDWLAADFKR 180
>AV412686
Length = 166
Score = 74.3 bits (181), Expect = 2e-14
Identities = 35/55 (63%), Positives = 44/55 (79%)
Frame = +2
Query: 230 GDTHLGGEDFDNRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTA 284
GDT+LGG DFDN +VN V F+RK+ KDI+ N ++LRRLR ACE+AKR LSST+
Sbjct: 2 GDTYLGGVDFDNNLVNFLVDFFQRKHNKDITENVKSLRRLRLACEKAKRILSSTS 166
>AV428240
Length = 342
Score = 60.8 bits (146), Expect = 3e-10
Identities = 31/80 (38%), Positives = 46/80 (56%)
Frame = +2
Query: 214 SLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFVQEFKRKNKKDISGNPRALRRLRTAC 273
S+ E G ++ + A D LGG DFD + HF ++FK + D+ N RA RLR AC
Sbjct: 2 SIAAFEFGQVKILSHAFDMSLGGRDFDEVLFCHFAEKFKEQYGIDVYSNVRACIRLRAAC 181
Query: 274 ERAKRTLSSTAQTTIEIDSL 293
E+ K+ LS+ A+ + I+ L
Sbjct: 182 EKLKKVLSANAEAPLTIECL 241
>BP064898
Length = 542
Score = 52.8 bits (125), Expect = 7e-08
Identities = 26/66 (39%), Positives = 38/66 (57%)
Frame = +1
Query: 74 KRLIGRRISDASVQSDMKLWPFKVIAGPGEKPMIGVNYKGEDKQFAAEEISSMVLIKMRE 133
KRLIG + SD +Q D+K PF V GP P+I Y GE + F ++ +M+L ++E
Sbjct: 337 KRLIGIQFSDPQLQRDLKSLPFLVTEGPDGYPLIHARYLGETRTFTPTQVFAMMLSNLKE 516
Query: 134 IAEAYL 139
I + L
Sbjct: 517 IXQKNL 534
>AV409958
Length = 389
Score = 34.7 bits (78), Expect = 0.019
Identities = 18/48 (37%), Positives = 27/48 (55%)
Frame = +2
Query: 243 MVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEI 290
+V+ +FKR D+ + +AL+RL E+AK LSS QT I +
Sbjct: 5 IVDWLAADFKRNEGIDLLKDKQALQRLTETAEKAKMELSSLTQTNISL 148
>AV427312
Length = 164
Score = 30.0 bits (66), Expect = 0.48
Identities = 14/26 (53%), Positives = 18/26 (68%)
Frame = +2
Query: 268 RLRTACERAKRTLSSTAQTTIEIDSL 293
+LR E+AKR LSS Q +EI+SL
Sbjct: 5 KLRREVEKAKRALSSQTQVRVEIESL 82
>TC9540
Length = 817
Score = 25.8 bits (55), Expect = 9.1
Identities = 13/34 (38%), Positives = 16/34 (46%)
Frame = -3
Query: 9 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTP 42
+IG+ VG WQ+D IA Q N TP
Sbjct: 308 SIGVATKLASITVGAWQYDPSNAIAIAQRNTVTP 207
>TC10576 similar to UP|Q93ZQ3 (Q93ZQ3) AT3g63200/F16M2_50, partial (39%)
Length = 751
Score = 25.8 bits (55), Expect = 9.1
Identities = 13/38 (34%), Positives = 22/38 (57%), Gaps = 3/38 (7%)
Frame = +2
Query: 176 IINEPTAAAIAYGLDKK---ATSVGEKNVLIFDLGGGT 210
++N PTAAA+ + L K G +++L+ LG G+
Sbjct: 119 VMNNPTAAAVTHVLHNKRDFPAVNGVEDLLVLSLGNGS 232
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.134 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,459,395
Number of Sequences: 28460
Number of extensions: 33973
Number of successful extensions: 157
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 152
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 152
length of query: 293
length of database: 4,897,600
effective HSP length: 90
effective length of query: 203
effective length of database: 2,336,200
effective search space: 474248600
effective search space used: 474248600
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 55 (25.8 bits)
Medicago: description of AC136140.11


