

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135848.10 - phase: 0
(346 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
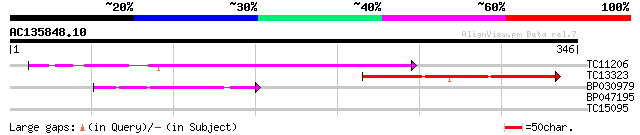
Score E
Sequences producing significant alignments: (bits) Value
TC11206 similar to UP|Q84V88 (Q84V88) Cyclin D, partial (43%) 141 2e-34
TC13323 weakly similar to UP|P93103 (P93103) Cyclin-D like prote... 105 1e-23
BP030979 41 3e-04
BP047195 31 0.27
TC15095 similar to UP|AAQ96377 (AAQ96377) Miraculin-like protein... 27 5.0
>TC11206 similar to UP|Q84V88 (Q84V88) Cyclin D, partial (43%)
Length = 816
Score = 141 bits (355), Expect = 2e-34
Identities = 87/241 (36%), Positives = 133/241 (55%), Gaps = 4/241 (1%)
Frame = +2
Query: 12 LLCSENNSTCFDDVVVDDSGISPSWDHTNVNLDNVGSDSFLCFVAQSEEIVKVMV-EKEK 70
LLC E ++ DD ++ + HT + D + F + +E+V ++ E+ +
Sbjct: 146 LLCEEQHTFDDDD---EEKATHATECHT------INDDPLILFWEEDDELVSLISNERGE 298
Query: 71 DHLPREDYLIRLRGGDLDL---SVRREALDWIWKAHAYYGFGPLSLCLSVNYLDRFLSVF 127
HL G LDL R E + WI K A++GF L+ L+V+Y DRF++
Sbjct: 299 THL-----------GSLDLHDGGTRVEGVAWISKVSAHFGFSALTTVLAVSYFDRFITSP 445
Query: 128 QFPRGVTWTVQLLAVACFSLAAKMEEVKVPQSVDLQVGEPKFVFQAKTIQRMELMILSSL 187
+F R W QL AVAC SLAAK+EE VP +DLQV E +FVF+AKTIQRMEL++LS+L
Sbjct: 446 RFQRDKPWMTQLSAVACLSLAAKVEETHVPLLLDLQVEESRFVFEAKTIQRMELLVLSTL 625
Query: 188 GWKMRALTPCSFIDYFLAKISCEKYPDKSLIARSVQLILNIIKGIDFLEFRSSEIAAAVA 247
W+M +TP SF ++ + ++ + + R +++LN+I + + S +AAA
Sbjct: 626 KWRMHPVTPISFFEHIIRRLGLKTRLHWEFLWRCERVLLNVIPDSRVMSYLPSTLAAAAM 805
Query: 248 I 248
I
Sbjct: 806 I 808
>TC13323 weakly similar to UP|P93103 (P93103) Cyclin-D like protein, partial
(19%)
Length = 514
Score = 105 bits (261), Expect = 1e-23
Identities = 66/124 (53%), Positives = 84/124 (67%), Gaps = 3/124 (2%)
Frame = +1
Query: 216 SLIARSVQLILNIIKGIDFLEFRSSEIAAAVAISLKELPTQEVDKAITDFFI---VDKER 272
SLI+RS QLIL+ I+G+DFL F+ SEIAAAVA+ + TQ VD T + V+KER
Sbjct: 22 SLISRSTQLILSTIRGLDFLAFKPSEIAAAVAMYVVG-ETQSVDTDQTISILIQHVEKER 198
Query: 273 VLKCVELIRDLSLIKVGGNNFASFVPQSPIGVLDAGCMSFKSDELTNGSCPNSSHSSPNA 332
++KCVE+I +LS K + A VP SPIGVL+AGC S+KSD TN SS +SP+A
Sbjct: 199 LMKCVEMIHELSSNKDSSAS-APCVPPSPIGVLEAGCFSYKSDVDTNAGSSCSSDNSPSA 375
Query: 333 KRMK 336
KR K
Sbjct: 376 KRRK 387
>BP030979
Length = 510
Score = 41.2 bits (95), Expect = 3e-04
Identities = 31/102 (30%), Positives = 54/102 (52%)
Frame = +3
Query: 52 LCFVAQSEEIVKVMVEKEKDHLPREDYLIRLRGGDLDLSVRREALDWIWKAHAYYGFGPL 111
L +V+ E ++ M E + P DY+ +++ D++ ++R +DW+ + Y
Sbjct: 219 LPYVSDIYEYLRSM-EVDPSKRPLPDYVQKVQR-DVNANMRGVXVDWLVEVSEEYKLVSD 392
Query: 112 SLCLSVNYLDRFLSVFQFPRGVTWTVQLLAVACFSLAAKMEE 153
+L V+Y+DRFLS+ R +QLL VA +A+K EE
Sbjct: 393 TLYFCVSYIDRFLSLNSLSR---QKLQLLGVASMLVASKYEE 509
>BP047195
Length = 390
Score = 31.2 bits (69), Expect = 0.27
Identities = 32/107 (29%), Positives = 51/107 (46%), Gaps = 6/107 (5%)
Frame = -2
Query: 229 IKGIDFLEFRSSEIAAAVAISL-KELPTQEVDK---AITDFFIVDKERVLKCVELIRDLS 284
I + FL +RSS IAAA +S E+P K A + + KE+++ C +LI++L
Sbjct: 389 IHDVSFLAYRSSCIAAAPILSASNEIPEWSFVKPEHAESWCQGLRKEKIIVCYKLIQELI 210
Query: 285 LIKVGGNNFASFVPQSPIGVLDA--GCMSFKSDELTNGSCPNSSHSS 329
+I +PQ + C+S L++ S +SS SS
Sbjct: 209 VINHSSGRTPKVLPQLRVTTRTRRWSCIS----SLSSSSSSSSSSSS 81
>TC15095 similar to UP|AAQ96377 (AAQ96377) Miraculin-like protein
(Fragment), partial (42%)
Length = 979
Score = 26.9 bits (58), Expect = 5.0
Identities = 14/45 (31%), Positives = 21/45 (46%)
Frame = +3
Query: 257 EVDKAITDFFIVDKERVLKCVELIRDLSLIKVGGNNFASFVPQSP 301
+++KA D+ V KC L R+L + GG+ S Q P
Sbjct: 648 KIEKADKDYVFSFCPSVCKCQTLCRELGIYDYGGDKHLSLSDQVP 782
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.136 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,975,446
Number of Sequences: 28460
Number of extensions: 79294
Number of successful extensions: 389
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 382
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 387
length of query: 346
length of database: 4,897,600
effective HSP length: 91
effective length of query: 255
effective length of database: 2,307,740
effective search space: 588473700
effective search space used: 588473700
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 56 (26.2 bits)
Medicago: description of AC135848.10


