

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135799.6 + phase: 0
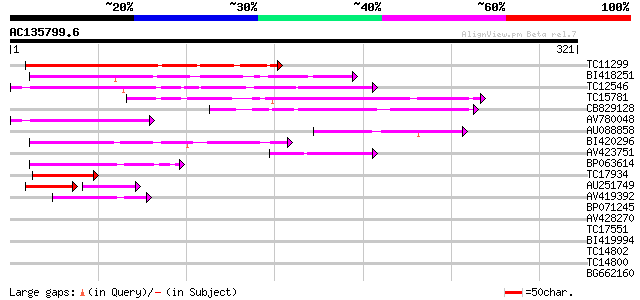
(321 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC11299 100 6e-22
BI418251 96 6e-21
TC12546 weakly similar to UP|Q9FJ30 (Q9FJ30) Similarity to heat ... 91 3e-19
TC15781 90 6e-19
CB829128 76 9e-15
AV780048 68 2e-12
AU088858 53 6e-08
BI420296 49 1e-06
AV423751 48 2e-06
BP063614 47 4e-06
TC17934 similar to PIR|T06667|T06667 argininosuccinate synthase ... 43 8e-05
AU251749 31 3e-04
AV419392 40 5e-04
BP071245 39 0.001
AV428270 34 0.029
TC17551 weakly similar to UP|AAR24662 (AAR24662) At5g26960, part... 34 0.038
BI419994 32 0.14
TC14802 weakly similar to UP|CAD56661 (CAD56661) S locus F-box (... 31 0.25
TC14800 weakly similar to UP|Q9FHP3 (Q9FHP3) Genomic DNA, chromo... 31 0.25
BG662160 31 0.32
>TC11299
Length = 498
Score = 99.8 bits (247), Expect = 6e-22
Identities = 64/147 (43%), Positives = 90/147 (60%), Gaps = 2/147 (1%)
Frame = +1
Query: 10 VIDRISSFPDDILIRILSSLPIEQACVTSILSKRWTHLWCFVPDLDFTKTKWKDQESYS- 68
++DRIS+ PD+I+ ILS LP E A T++LSKRWTHLW V LDF+ + + + +
Sbjct: 79 MVDRISALPDEIICHILSFLPTENAVATAVLSKRWTHLWRSVSALDFSSVRMYEPDDHRF 258
Query: 69 RFEEFVFSVLYSREAAGNHSINTFILDIEYDSSELLLDGIKNIHKWVDIVVKSKVQNIHL 128
F + V+SVL R AA I F L+++ D + LD I NIHKWV+ V + +V I
Sbjct: 259 FFSDIVYSVLLFRNAA--TPIKKFSLELDDDVAVANLD-IANIHKWVNFVTQCRVGGIE- 426
Query: 129 YPHVPLNGEETI-LPKFPMSSILSCTT 154
H+ L+ + I LP+ P+ SILSC T
Sbjct: 427 --HLRLHFMDFIQLPQLPI-SILSCKT 498
>BI418251
Length = 546
Score = 96.3 bits (238), Expect = 6e-21
Identities = 70/189 (37%), Positives = 99/189 (52%), Gaps = 3/189 (1%)
Frame = +1
Query: 12 DRISSFPDDILIRILSSLPIEQACVTSILSKRWTHLWCFVPDLDFTK---TKWKDQESYS 68
DRIS PD++L +ILS LP E A TS+L KRW+ LW VP LDF K K + S
Sbjct: 37 DRISKLPDEVLCQILSFLPTEDAVATSVLCKRWSSLWLSVPTLDFDDYRYLKGKKLKLQS 216
Query: 69 RFEEFVFSVLYSREAAGNHSINTFILDIEYDSSELLLDGIKNIHKWVDIVVKSKVQNIHL 128
F FV++++ SR A + I F L + + ++ W++ ++ +V+N+ +
Sbjct: 217 SFINFVYAIILSR--ALHQPIKNFTLSVISEECP-----YPDVKVWLNAAMQRQVENLDI 375
Query: 129 YPHVPLNGEETILPKFPMSSILSCTTLVVLKLRWFNLTVVSDLSIRLPSLKTLHLKEIYF 188
+ + LP SILSCTTLVVLKL V S + LPSLKTLHL+ +
Sbjct: 376 CLSL------STLP----CSILSCTTLVVLKLSDVKFHVFS--CVDLPSLKTLHLELVVI 519
Query: 189 DQQRDFMML 197
+ M L
Sbjct: 520 LNPQSLMDL 546
>TC12546 weakly similar to UP|Q9FJ30 (Q9FJ30) Similarity to heat shock
transcription factor HSF30, partial (9%)
Length = 755
Score = 90.9 bits (224), Expect = 3e-19
Identities = 74/219 (33%), Positives = 110/219 (49%), Gaps = 11/219 (5%)
Frame = +2
Query: 1 MSKQKISTSVIDRISSFPDDILIRILSSLPIEQACVTSILSKRWTHLWCFVPDLDFTKTK 60
M K K+ D IS+ PD +L ILS L ++A TS+LS+RW LW VP L F
Sbjct: 128 MKKMKME----DGISTLPDALLCHILSFLTSKEAVATSVLSRRWIPLWRSVPTLHFKDAN 295
Query: 61 WKD----------QESYSRFEEFVFSVLYSREAAGNHSINTFILDIEYDSSELLLDGIKN 110
+ ++ SR + V++V+ SR+ I F L + D + D N
Sbjct: 296 YHTDIGHADHDIVKDVRSRHVQSVYAVILSRDF--QLPIKKFYLRLN-DVCQPFYDP-AN 463
Query: 111 IHKWVDIVVKSKVQNIHLYPHVPLNGEETILPKFPMSSILSCTTLVVLKLRWFNLTVVSD 170
+ WV+ VV+ +++++ + P+ P+ +SSI SC TLVVLKLR L +
Sbjct: 464 VSVWVNAVVQRQLEHLDISLPYPM----LSTPRANLSSIFSCRTLVVLKLRG-GLELKRF 628
Query: 171 LSIRLPSLKTLHLKEIYFDQQRDFMM-LLDGCPVLEDLQ 208
S+ P LK LHL+ ++ LL GCPVLE+L+
Sbjct: 629 PSVHFPCLKVLHLQGALLLHDVPYLAELLSGCPVLENLK 745
>TC15781
Length = 621
Score = 89.7 bits (221), Expect = 6e-19
Identities = 71/208 (34%), Positives = 104/208 (49%), Gaps = 5/208 (2%)
Frame = +2
Query: 67 YSRFEEFVFSVLYSREAAGNHSINTFILDIEYDSSELLLDGIKNIHKWVDIVVKSKVQNI 126
Y E FV + + +R+ I F L E S + L+G +H W++ V+ +++N+
Sbjct: 5 YPFLENFVNATILARDE--RQPITRF*LLYE---STVSLEGSDLVHDWLNTVMPRRIENL 169
Query: 127 HLYPHVPLNGEETILPKFPMS-----SILSCTTLVVLKLRWFNLTVVSDLSIRLPSLKTL 181
E +P P+S I SC TLVVLKL+ + L V + LPSLK+L
Sbjct: 170 -----------EICIP--PLSRGIGLKIYSCRTLVVLKLKGYPLGVYVSSHVELPSLKSL 310
Query: 182 HLKEIYFDQQRDFMMLLDGCPVLEDLQLCYIYMTRQSHHSLDDFESSSMLKKLNRADITD 241
HL++++F + F+ L+ GCP+LEDL+ + S F S L KL I D
Sbjct: 311 HLEKVHFVDFQSFLKLICGCPLLEDLEAGTVSYLDTSFDQEFRFRS---LPKLVSVVIND 481
Query: 242 CECYFPVKSLSNLEFLRIKLSEVCFIIE 269
FP+K +SN EFL I+ EVC E
Sbjct: 482 YNFRFPLKIISNAEFLSIE--EVCIYPE 559
>CB829128
Length = 463
Score = 75.9 bits (185), Expect = 9e-15
Identities = 59/154 (38%), Positives = 84/154 (54%), Gaps = 2/154 (1%)
Frame = +3
Query: 114 WVDIVVKSKVQNIHLYPHVPLNGEETILPKFPMSSILSCTTLVVLKLRWFNLTVVSDLSI 173
W++ ++ +V+N+ + E + P S IL CTTLV+LKL ++T + S+
Sbjct: 33 WLNAAIQRQVENLEI---------ELTDSQMPCS-ILRCTTLVILKLD--SVTFDAFTSV 176
Query: 174 RLPSLKTLHLKEIYFDQQRDFMMLLDGCPVLEDLQLCYIYMTRQSHHSLDDFESS-SMLK 232
LPSLKTLHL E+ + + + LL GCP+LEDL+ Q + FE L
Sbjct: 177 YLPSLKTLHLFEVDLVKAQCLIDLLYGCPILEDLK-------TQIYFRDGSFEGKVKTLS 335
Query: 233 KLNRADI-TDCECYFPVKSLSNLEFLRIKLSEVC 265
KL RAD+ D PVK+ N+EFLRI+ EVC
Sbjct: 336 KLVRADVFLDGHFIIPVKAFQNVEFLRIE--EVC 431
>AV780048
Length = 501
Score = 67.8 bits (164), Expect = 2e-12
Identities = 39/83 (46%), Positives = 48/83 (56%), Gaps = 1/83 (1%)
Frame = +2
Query: 1 MSKQKISTSVIDRISSFPDDILIRILSSLPIEQACVTSILSKRWTHLWCFVPDLDFTKTK 60
M K K+ DR SS PD IL ILS L ++A TSILSKRW LW VP LDF +
Sbjct: 230 MKKMKME----DRFSSLPDPILCHILSLLTTKEAVTTSILSKRWIPLWRSVPTLDFDDSN 397
Query: 61 W-KDQESYSRFEEFVFSVLYSRE 82
W E Y+R E V+ V+ +R+
Sbjct: 398 WYGGDEVYARLVEAVYKVILARD 466
>AU088858
Length = 765
Score = 53.1 bits (126), Expect = 6e-08
Identities = 33/89 (37%), Positives = 51/89 (57%), Gaps = 2/89 (2%)
Frame = +3
Query: 173 IRLPSLKTLHLKEIYFDQQRDFMMLLDGCPVLEDLQLCYIYMTRQSHHSLDDFESSSM-- 230
+ LPSLKTL L + F + + M LL GCP+LE Y++++R + ++++
Sbjct: 15 VHLPSLKTLQLYCLIFPEPQYVMELLSGCPILE-----YMHLSRMRYADCSSPSNTNVKS 179
Query: 231 LKKLNRADITDCECYFPVKSLSNLEFLRI 259
L KL ADI+ P+K + N+EFLRI
Sbjct: 180 LTKLVSADISFIGFEIPLKVICNVEFLRI 266
>BI420296
Length = 592
Score = 48.5 bits (114), Expect = 1e-06
Identities = 46/152 (30%), Positives = 68/152 (44%), Gaps = 3/152 (1%)
Frame = +2
Query: 12 DRISSFPDDILIRILSSLPIEQACVTSILSKRWTHLWCFVPDLDFTKTKWKDQESYSRFE 71
DR+S PD +L+ +L + +QA T +LS RW LW V L + D + RF
Sbjct: 194 DRLSDLPDLVLLHVLKFMSTKQAVQTCVLSTRWKDLWKGVTTLALNSS---DFATAPRFS 364
Query: 72 EFVFSVLYSREAAGNHSINTFILDIEYD---SSELLLDGIKNIHKWVDIVVKSKVQNIHL 128
EF+ VL R N S++ LD+ ELL ++ + V VQ++ +
Sbjct: 365 EFLSCVLSHR----NDSVSLHNLDLRRKGCVEPELL-------NRVMSYAVSHDVQSLTI 511
Query: 129 YPHVPLNGEETILPKFPMSSILSCTTLVVLKL 160
++ L + P I SC TL LKL
Sbjct: 512 EFNLYLKLGFKLHP-----CIFSCRTLTYLKL 592
>AV423751
Length = 254
Score = 48.1 bits (113), Expect = 2e-06
Identities = 27/61 (44%), Positives = 36/61 (58%)
Frame = +2
Query: 148 SILSCTTLVVLKLRWFNLTVVSDLSIRLPSLKTLHLKEIYFDQQRDFMMLLDGCPVLEDL 207
S+ SCTTLVVLKL L + S+ LP LK LHL+++ + +L GC LEDL
Sbjct: 74 SMFSCTTLVVLKLEGLELKA-NFSSVDLPFLKVLHLQDLLLQDEGCLAEILSGCLALEDL 250
Query: 208 Q 208
+
Sbjct: 251 K 253
>BP063614
Length = 508
Score = 47.0 bits (110), Expect = 4e-06
Identities = 32/88 (36%), Positives = 48/88 (54%)
Frame = +2
Query: 12 DRISSFPDDILIRILSSLPIEQACVTSILSKRWTHLWCFVPDLDFTKTKWKDQESYSRFE 71
DR+S PD IL+ ILS + + A T ILS RW LW +P L + + +Y F
Sbjct: 167 DRLSDLPDCILLHILSFVKAKAAVQTCILSTRWKDLWKRLPSLILLSSNF---WTYKSFT 337
Query: 72 EFVFSVLYSREAAGNHSINTFILDIEYD 99
+FV +L R+ G+ +++ LD E+D
Sbjct: 338 KFVSRLLTLRD--GSTALHG--LDFEHD 409
>TC17934 similar to PIR|T06667|T06667 argininosuccinate synthase -
Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (11%)
Length = 564
Score = 42.7 bits (99), Expect = 8e-05
Identities = 19/37 (51%), Positives = 25/37 (67%)
Frame = -1
Query: 14 ISSFPDDILIRILSSLPIEQACVTSILSKRWTHLWCF 50
I+ PD I ILS LPI +A TSILS++W ++W F
Sbjct: 564 INQLPDGIPGAILSKLPINEAAKTSILSRKWRYMWTF 454
>AU251749
Length = 362
Score = 30.8 bits (68), Expect(2) = 3e-04
Identities = 14/34 (41%), Positives = 20/34 (58%), Gaps = 1/34 (2%)
Frame = +1
Query: 42 KRWTHLWCFVPDLDFTKTK-WKDQESYSRFEEFV 74
KRW LW VP D + W+D+E+Y+R + V
Sbjct: 214 KRWKPLWRSVPTRDLDDGRFWRDREAYARLVQGV 315
Score = 29.3 bits (64), Expect(2) = 3e-04
Identities = 15/29 (51%), Positives = 19/29 (64%)
Frame = +2
Query: 10 VIDRISSFPDDILIRILSSLPIEQACVTS 38
++D IS+ PD IL ILS LP +Q TS
Sbjct: 131 MVDWISALPDTILNFILSFLPTKQVVATS 217
>AV419392
Length = 318
Score = 40.0 bits (92), Expect = 5e-04
Identities = 23/56 (41%), Positives = 30/56 (53%)
Frame = +3
Query: 25 ILSSLPIEQACVTSILSKRWTHLWCFVPDLDFTKTKWKDQESYSRFEEFVFSVLYS 80
ILS + + A S+LSKRW LW +P L+F + YS F FV VL+S
Sbjct: 6 ILSYVETKDAMQCSVLSKRWRSLWRALPVLNFDSASF---THYSSFANFVLHVLHS 164
>BP071245
Length = 552
Score = 39.3 bits (90), Expect = 0.001
Identities = 19/43 (44%), Positives = 25/43 (57%)
Frame = +1
Query: 12 DRISSFPDDILIRILSSLPIEQACVTSILSKRWTHLWCFVPDL 54
DR+S PD +L+ ILS + + A T LS RW LW +P L
Sbjct: 382 DRLSDLPDCVLLHILSFVNAKYAVQTCTLSTRWKDLWKRLPSL 510
>AV428270
Length = 428
Score = 34.3 bits (77), Expect = 0.029
Identities = 26/92 (28%), Positives = 44/92 (47%)
Frame = +1
Query: 198 LDGCPVLEDLQLCYIYMTRQSHHSLDDFESSSMLKKLNRADITDCECYFPVKSLSNLEFL 257
L GCP+LE+L+ I++T +S + L KL RADI ++ N+ FL
Sbjct: 7 LAGCPILENLKARRIWITGESSFYRGGELADLGLPKLIRADIVRSRLPM-LQPFYNVAFL 183
Query: 258 RIKLSEVCFIIEWNESEQCDSFVFLIFNFIEL 289
R+ + EV + + + V ++N + L
Sbjct: 184 RVDIEEVHYAFPYFHNLTHLELVIELYNSLVL 279
>TC17551 weakly similar to UP|AAR24662 (AAR24662) At5g26960, partial (22%)
Length = 505
Score = 33.9 bits (76), Expect = 0.038
Identities = 14/34 (41%), Positives = 22/34 (64%)
Frame = +2
Query: 14 ISSFPDDILIRILSSLPIEQACVTSILSKRWTHL 47
ISS PDDI++ L+ +P S++ +RW+HL
Sbjct: 128 ISSLPDDIILDFLTRVPPSSLPSLSLVCRRWSHL 229
>BI419994
Length = 549
Score = 32.0 bits (71), Expect = 0.14
Identities = 16/53 (30%), Positives = 27/53 (50%)
Frame = +2
Query: 7 STSVIDRISSFPDDILIRILSSLPIEQACVTSILSKRWTHLWCFVPDLDFTKT 59
ST + S PD+++++IL LP+ + K W +L + D FTK+
Sbjct: 17 STESLSLFSHLPDELILQILLRLPVRSLLRFKTVCKSWRYL---ISDPQFTKS 166
>TC14802 weakly similar to UP|CAD56661 (CAD56661) S locus F-box (SLF)-S4
protein, partial (10%)
Length = 551
Score = 31.2 bits (69), Expect = 0.25
Identities = 15/41 (36%), Positives = 23/41 (55%)
Frame = +3
Query: 18 PDDILIRILSSLPIEQACVTSILSKRWTHLWCFVPDLDFTK 58
PD+++I ILS LP++ ++SK W F+ D F K
Sbjct: 24 PDELIIEILSWLPVKSLLQFRVVSKTWK---SFISDPQFVK 137
>TC14800 weakly similar to UP|Q9FHP3 (Q9FHP3) Genomic DNA, chromosome 5,
TAC clone:K14B20, partial (7%)
Length = 782
Score = 31.2 bits (69), Expect = 0.25
Identities = 15/41 (36%), Positives = 23/41 (55%)
Frame = +3
Query: 18 PDDILIRILSSLPIEQACVTSILSKRWTHLWCFVPDLDFTK 58
PD+++I ILS LP++ ++SK W F+ D F K
Sbjct: 51 PDELIIEILSWLPVKSLLQFRVVSKTWK---SFISDPQFVK 164
>BG662160
Length = 445
Score = 30.8 bits (68), Expect = 0.32
Identities = 20/87 (22%), Positives = 36/87 (40%)
Frame = +2
Query: 13 RISSFPDDILIRILSSLPIEQACVTSILSKRWTHLWCFVPDLDFTKTKWKDQESYSRFEE 72
R P+++ + ILS LP++ +SK W + + D F K S +R +
Sbjct: 11 RAQFLPEELRLEILSWLPVKSLVRFRCVSKSWKSI---ISDSQFIKLHLHRSSSTTRNTD 181
Query: 73 FVFSVLYSREAAGNHSINTFILDIEYD 99
F + + S +T + +YD
Sbjct: 182FAYLQSLITSPRKSRSASTIAIPDDYD 262
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.325 0.139 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,748,398
Number of Sequences: 28460
Number of extensions: 104221
Number of successful extensions: 864
Number of sequences better than 10.0: 56
Number of HSP's better than 10.0 without gapping: 850
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 853
length of query: 321
length of database: 4,897,600
effective HSP length: 90
effective length of query: 231
effective length of database: 2,336,200
effective search space: 539662200
effective search space used: 539662200
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 55 (25.8 bits)
Medicago: description of AC135799.6


