

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135799.5 + phase: 0
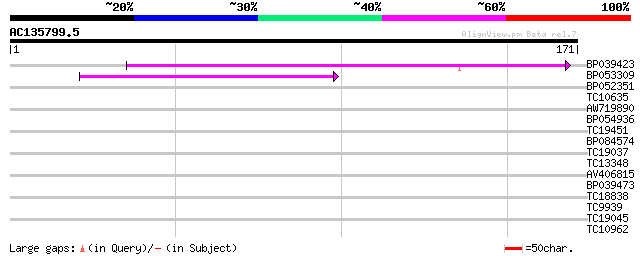
(171 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BP039423 50 2e-07
BP053309 42 4e-05
BP052351 32 0.058
TC10635 similar to UP|Q946U8 (Q946U8) Long cell-linked locus pro... 27 1.9
AW719890 27 2.4
BP054936 27 2.4
TC19451 similar to GB|AAM65667.1|21593700|AY088122 J8-like prote... 26 3.2
BP084574 26 4.2
TC19037 similar to PIR|S71851|S71851 heat shock transcription fa... 26 4.2
TC13348 weakly similar to UP|Q9LUJ4 (Q9LUJ4) Gb|AAF26800.1, part... 26 4.2
AV406815 25 5.4
BP039473 25 5.4
TC18838 weakly similar to PIR|PC4168|PC4168 pectinesterase 2 pr... 25 7.1
TC9939 homologue to UP|GSA_SOYBN (P45621) Glutamate-1-semialdehy... 25 7.1
TC19045 similar to GB|AAO64820.1|29028882|BT005885 At3g49990 {Ar... 25 9.3
TC10962 similar to UP|Q9SV20 (Q9SV20) Beta-COP-like protein, par... 25 9.3
>BP039423
Length = 566
Score = 50.1 bits (118), Expect = 2e-07
Identities = 37/135 (27%), Positives = 59/135 (43%), Gaps = 1/135 (0%)
Frame = +1
Query: 36 PTSIIFMNMLLVDEKGGRIHATTRKDLVAKFRSMVQEGGTYQLENAIVDFNESPYKVTSH 95
P+ +N++L+DE+G +I A +K ++KFR EG Y++ V N ++ H
Sbjct: 25 PSKPYAINLVLIDEEGVKIEAYIKKGYMSKFRGEFAEGNVYKITYFAVGDNGGSFRACEH 204
Query: 96 KHKLSMMHNSTFTKVHLPAIPMNVFEFKPFNEILSSTVE-EVSTDVIGHVIERGDIRETE 154
K+ + IP K EI + E + D IG + E R+ E
Sbjct: 205 AFKIFFTSRTRAVPDFSDVIPTVGLTLKNSAEIKGTLGESDFLIDYIGLLAELPSERQYE 384
Query: 155 KDRRKSRVIDLTLED 169
K +R I+L L D
Sbjct: 385 KSGNITRKIELELID 429
>BP053309
Length = 484
Score = 42.4 bits (98), Expect = 4e-05
Identities = 21/78 (26%), Positives = 40/78 (50%)
Frame = +2
Query: 22 VRVAHIWLIREKKVPTSIIFMNMLLVDEKGGRIHATTRKDLVAKFRSMVQEGGTYQLENA 81
VRV +W + P+ ++++L+D +G +I +K ++ KFR + EG Y++
Sbjct: 68 VRVLRVWDMFPVGEPSKPYAIHVVLIDVEGVKIEGIIKKGMLKKFRGELVEGSVYRITYF 247
Query: 82 IVDFNESPYKVTSHKHKL 99
V N ++ T H K+
Sbjct: 248 NVISNSGAFRATEHGFKI 301
>BP052351
Length = 517
Score = 32.0 bits (71), Expect = 0.058
Identities = 19/72 (26%), Positives = 31/72 (42%)
Frame = -3
Query: 72 EGGTYQLENAIVDFNESPYKVTSHKHKLSMMHNSTFTKVHLPAIPMNVFEFKPFNEILSS 131
EGG V +NE+ K S ++S N+++ K + A F FK +I S
Sbjct: 503 EGGDIDKXTRFVAYNETENKGNSEMSEISKTQNNSYVKENTGATKDEEFNFKGTTQISDS 324
Query: 132 TVEEVSTDVIGH 143
+ + D + H
Sbjct: 323 SSANGTFDSVEH 288
>TC10635 similar to UP|Q946U8 (Q946U8) Long cell-linked locus protein,
partial (16%)
Length = 1243
Score = 26.9 bits (58), Expect = 1.9
Identities = 14/35 (40%), Positives = 23/35 (65%), Gaps = 1/35 (2%)
Frame = +2
Query: 79 ENAIVDFNESPYKVTSHKHKLSMMHNSTF-TKVHL 112
E A DF+ + + T +HKL+M+ +STF ++HL
Sbjct: 896 EAAAPDFSSTIH*NTHERHKLNMLIHSTFKAQIHL 1000
>AW719890
Length = 599
Score = 26.6 bits (57), Expect = 2.4
Identities = 17/59 (28%), Positives = 29/59 (48%), Gaps = 9/59 (15%)
Frame = +1
Query: 109 KVHLPAIPMNVFEFKPFNEILSSTVEEVS---------TDVIGHVIERGDIRETEKDRR 158
K+ LPA+P + F +P +++ TVE S D +G +I+ + E+ RR
Sbjct: 220 KLPLPAMPNSEFAIQPTKVLVNVTVENSSGAVKLVVLPEDTVGDLIKAALVFYKEEKRR 396
>BP054936
Length = 537
Score = 26.6 bits (57), Expect = 2.4
Identities = 20/73 (27%), Positives = 34/73 (46%), Gaps = 6/73 (8%)
Frame = +3
Query: 103 HNSTFTKVHLPAIPMNVFEFKPFNEILSSTV------EEVSTDVIGHVIERGDIRETEKD 156
H ST T +HLP PF + L++ + + DVIG ++ + ++ K
Sbjct: 108 HPSTIT*IHLPINAHTTLTPAPFTQPLAAEIALTMGQSDYLVDVIG-LVGVSEEKQYLKS 284
Query: 157 RRKSRVIDLTLED 169
+ +RVI + L D
Sbjct: 285 FKVTRVIHIELTD 323
>TC19451 similar to GB|AAM65667.1|21593700|AY088122 J8-like protein
{Arabidopsis thaliana;}, partial (35%)
Length = 480
Score = 26.2 bits (56), Expect = 3.2
Identities = 10/23 (43%), Positives = 17/23 (73%)
Frame = -3
Query: 24 VAHIWLIREKKVPTSIIFMNMLL 46
+AH+W++ + KVP S F+N+ L
Sbjct: 328 LAHVWMVLQSKVPES--FLNLSL 266
>BP084574
Length = 336
Score = 25.8 bits (55), Expect = 4.2
Identities = 14/43 (32%), Positives = 21/43 (48%)
Frame = +1
Query: 57 TTRKDLVAKFRSMVQEGGTYQLENAIVDFNESPYKVTSHKHKL 99
TT + + K+ +M Q G L N + +FN V + HKL
Sbjct: 7 TTSRFFLCKYWTMFQ*GRAESLSNEVWNFNSCKIFVEAIAHKL 135
>TC19037 similar to PIR|S71851|S71851 heat shock transcription factor HSF4 -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(5%)
Length = 364
Score = 25.8 bits (55), Expect = 4.2
Identities = 11/29 (37%), Positives = 17/29 (57%)
Frame = -3
Query: 88 SPYKVTSHKHKLSMMHNSTFTKVHLPAIP 116
SP+ T + K S +H+ST T + P +P
Sbjct: 92 SPFSHTPNNFKFSPLHSSTTTSLLDPLVP 6
>TC13348 weakly similar to UP|Q9LUJ4 (Q9LUJ4) Gb|AAF26800.1, partial (7%)
Length = 539
Score = 25.8 bits (55), Expect = 4.2
Identities = 11/28 (39%), Positives = 18/28 (64%)
Frame = -2
Query: 88 SPYKVTSHKHKLSMMHNSTFTKVHLPAI 115
+P ++ S K SM+ STFT+ H+P +
Sbjct: 343 NPREINSSKE*ASMIKFSTFTQGHVPKV 260
>AV406815
Length = 426
Score = 25.4 bits (54), Expect = 5.4
Identities = 14/36 (38%), Positives = 18/36 (49%), Gaps = 4/36 (11%)
Frame = +2
Query: 73 GGTYQLEN----AIVDFNESPYKVTSHKHKLSMMHN 104
G T Q+ N AI N PY ++ H H L M+ N
Sbjct: 122 GPTVQVRNGDSVAIKVTNAGPYNISIHWHGLRMLRN 229
>BP039473
Length = 501
Score = 25.4 bits (54), Expect = 5.4
Identities = 12/30 (40%), Positives = 18/30 (60%)
Frame = +3
Query: 88 SPYKVTSHKHKLSMMHNSTFTKVHLPAIPM 117
+PYK+T++ L +HNST K + PM
Sbjct: 138 NPYKITTNSTILGTIHNSTQFKSD*FSFPM 227
>TC18838 weakly similar to PIR|PC4168|PC4168 pectinesterase 2 precursor -
Arabidopsis thaliana (fragment)
{Arabidopsis thaliana;} , partial (9%)
Length = 488
Score = 25.0 bits (53), Expect = 7.1
Identities = 11/21 (52%), Positives = 15/21 (71%)
Frame = +3
Query: 90 YKVTSHKHKLSMMHNSTFTKV 110
+KVTSHK +S++ N TF V
Sbjct: 351 HKVTSHKDVISLVLNITFRAV 413
>TC9939 homologue to UP|GSA_SOYBN (P45621) Glutamate-1-semialdehyde
2,1-aminomutase, chloroplast precursor (GSA)
(Glutamate-1-semialdehyde aminotransferase) (GSA-AT) ,
partial (52%)
Length = 903
Score = 25.0 bits (53), Expect = 7.1
Identities = 13/40 (32%), Positives = 20/40 (49%)
Frame = -2
Query: 15 RKNLKMCVRVAHIWLIREKKVPTSIIFMNMLLVDEKGGRI 54
RK L+ C W ++K P +++VDEKGG +
Sbjct: 242 RKLLQTCEE*VWCWSWGQRKRPL*WKEFELVVVDEKGGEV 123
>TC19045 similar to GB|AAO64820.1|29028882|BT005885 At3g49990 {Arabidopsis
thaliana;}, partial (11%)
Length = 874
Score = 24.6 bits (52), Expect = 9.3
Identities = 9/42 (21%), Positives = 25/42 (59%)
Frame = +3
Query: 9 SAVSGGRKNLKMCVRVAHIWLIREKKVPTSIIFMNMLLVDEK 50
S+++ G+ + +C +W+ ++ PT++ F L++D++
Sbjct: 309 SSMT*GKA*ISICFCRMSVWICFLEQSPTTVSFCFCLIIDQR 434
>TC10962 similar to UP|Q9SV20 (Q9SV20) Beta-COP-like protein, partial (36%)
Length = 1068
Score = 24.6 bits (52), Expect = 9.3
Identities = 10/19 (52%), Positives = 12/19 (62%)
Frame = -3
Query: 85 FNESPYKVTSHKHKLSMMH 103
F P KVTS+ H+LS H
Sbjct: 217 FRSQPLKVTSNTHQLSTEH 161
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.134 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,391,053
Number of Sequences: 28460
Number of extensions: 27738
Number of successful extensions: 168
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 168
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 168
length of query: 171
length of database: 4,897,600
effective HSP length: 84
effective length of query: 87
effective length of database: 2,506,960
effective search space: 218105520
effective search space used: 218105520
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 52 (24.6 bits)
Medicago: description of AC135799.5


