

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135798.2 + phase: 0
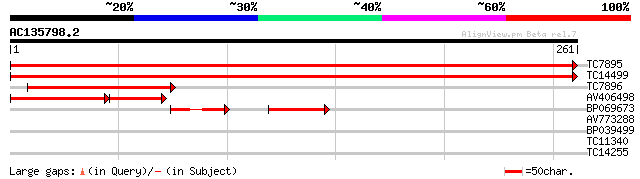
(261 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC7895 homologue to UP|Q8GTE3 (Q8GTE3) Ribosomal protein S3a, co... 503 e-143
TC14499 homologue to UP|Q8GTE3 (Q8GTE3) Ribosomal protein S3a, c... 487 e-139
TC7896 homologue to UP|Q8GTE3 (Q8GTE3) Ribosomal protein S3a, pa... 125 7e-30
AV406498 80 1e-25
BP069673 52 8e-13
AV773288 38 0.002
BP039499 30 0.55
TC11340 27 3.5
TC14255 26 7.9
>TC7895 homologue to UP|Q8GTE3 (Q8GTE3) Ribosomal protein S3a, complete
Length = 1106
Score = 503 bits (1296), Expect = e-143
Identities = 251/261 (96%), Positives = 257/261 (98%)
Frame = +3
Query: 1 MAVGKNKRISKGKKGGKKKAADPFAKKDWYDIKAPSVFQVKNVGKTLVSRTQGTKIASEG 60
MAVGKNKRISKGKKG KKKAADPFAKKDWYDIKAPS+FQVKNVGKTLVSRTQGTKIASEG
Sbjct: 63 MAVGKNKRISKGKKGSKKKAADPFAKKDWYDIKAPSLFQVKNVGKTLVSRTQGTKIASEG 242
Query: 61 LKHRVFEVSLADLQGDEDNAFRKIRLRAEDVQGKNLLTNFWGMDFTTDKLRSLVRKWQTL 120
LKHRVFEVSLADLQGDEDNAFRKIRLRAEDVQGKN+LTNFWGMDFTTDKLRSLVRKWQTL
Sbjct: 243 LKHRVFEVSLADLQGDEDNAFRKIRLRAEDVQGKNVLTNFWGMDFTTDKLRSLVRKWQTL 422
Query: 121 IEAHVDVKTTDNYTLRMFCIGFTKRRSNQVKRTCYAQSSQIRQIRRKMREIMINQATSCD 180
IEAHVDVKTTDNYTLRMFCI FTKRRSNQVKRTCYAQSSQIRQIRRKMREIMINQATSCD
Sbjct: 423 IEAHVDVKTTDNYTLRMFCIAFTKRRSNQVKRTCYAQSSQIRQIRRKMREIMINQATSCD 602
Query: 181 LKDLVRKFIPEMIGKEIEKATSSIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDYSEDVG 240
LK+LVRKFIPEMIGKEIEKATSSIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDYSEDVG
Sbjct: 603 LKELVRKFIPEMIGKEIEKATSSIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDYSEDVG 782
Query: 241 TKVERPADETMVEGTPEIVGA 261
TKV+RPADET+VEG E+VGA
Sbjct: 783 TKVDRPADETVVEGATEVVGA 845
>TC14499 homologue to UP|Q8GTE3 (Q8GTE3) Ribosomal protein S3a, complete
Length = 1119
Score = 487 bits (1254), Expect = e-139
Identities = 241/261 (92%), Positives = 253/261 (96%)
Frame = +2
Query: 1 MAVGKNKRISKGKKGGKKKAADPFAKKDWYDIKAPSVFQVKNVGKTLVSRTQGTKIASEG 60
MAVGKNKRISKGKKG KKKA+DPFAKKDWYDIKAPSVFQVKNVGKTLVSRTQGTKIASEG
Sbjct: 53 MAVGKNKRISKGKKGSKKKASDPFAKKDWYDIKAPSVFQVKNVGKTLVSRTQGTKIASEG 232
Query: 61 LKHRVFEVSLADLQGDEDNAFRKIRLRAEDVQGKNLLTNFWGMDFTTDKLRSLVRKWQTL 120
LKHRVFEVSLADLQGDE+ AFRKIRLRAEDVQGKN+LTNFWGM+FTTDKLRSLVRKWQTL
Sbjct: 233 LKHRVFEVSLADLQGDEEQAFRKIRLRAEDVQGKNVLTNFWGMNFTTDKLRSLVRKWQTL 412
Query: 121 IEAHVDVKTTDNYTLRMFCIGFTKRRSNQVKRTCYAQSSQIRQIRRKMREIMINQATSCD 180
IEAHVDVKTTDNYTLRMFCIGFTKRR+NQVKRTCYAQSSQIRQIRRKMREIM NQATSCD
Sbjct: 413 IEAHVDVKTTDNYTLRMFCIGFTKRRANQVKRTCYAQSSQIRQIRRKMREIMTNQATSCD 592
Query: 181 LKDLVRKFIPEMIGKEIEKATSSIYPLQNVFIRKVKILKAPKFDLGKLMEVHGDYSEDVG 240
LK+LVRKFIPE+IGKE+EKAT+ IYPLQNVF+RKVKILKAPKFDLGKLMEVHGDYSEDVG
Sbjct: 593 LKELVRKFIPEVIGKEVEKATTGIYPLQNVFVRKVKILKAPKFDLGKLMEVHGDYSEDVG 772
Query: 241 TKVERPADETMVEGTPEIVGA 261
TKV+RPADET+ E EIVGA
Sbjct: 773 TKVDRPADETVAEEPTEIVGA 835
>TC7896 homologue to UP|Q8GTE3 (Q8GTE3) Ribosomal protein S3a, partial
(25%)
Length = 592
Score = 125 bits (314), Expect = 7e-30
Identities = 63/68 (92%), Positives = 64/68 (93%)
Frame = +3
Query: 9 ISKGKKGGKKKAADPFAKKDWYDIKAPSVFQVKNVGKTLVSRTQGTKIASEGLKHRVFEV 68
ISKGKKG KKKAADPFAKKDWYDIKAPS+FQVKNVGKTLVSRTQGTKIASEGLKHRVFEV
Sbjct: 3 ISKGKKGSKKKAADPFAKKDWYDIKAPSLFQVKNVGKTLVSRTQGTKIASEGLKHRVFEV 182
Query: 69 SLADLQGD 76
SLA L D
Sbjct: 183SLAFLLHD 206
>AV406498
Length = 248
Score = 79.7 bits (195), Expect(2) = 1e-25
Identities = 36/46 (78%), Positives = 40/46 (86%)
Frame = +1
Query: 1 MAVGKNKRISKGKKGGKKKAADPFAKKDWYDIKAPSVFQVKNVGKT 46
+ VGKNKRISKGKKG KK+A DPFAKKDWY IKAP+V QVKNVG +
Sbjct: 34 ITVGKNKRISKGKKGSKKRACDPFAKKDWYGIKAPAVIQVKNVGNS 171
Score = 52.8 bits (125), Expect(2) = 1e-25
Identities = 26/26 (100%), Positives = 26/26 (100%)
Frame = +3
Query: 47 LVSRTQGTKIASEGLKHRVFEVSLAD 72
LVSRTQGTKIASEGLKHRVFEVSLAD
Sbjct: 171 LVSRTQGTKIASEGLKHRVFEVSLAD 248
>BP069673
Length = 516
Score = 51.6 bits (122), Expect(2) = 8e-13
Identities = 22/28 (78%), Positives = 26/28 (92%)
Frame = +2
Query: 120 LIEAHVDVKTTDNYTLRMFCIGFTKRRS 147
LI++HVDV TTDNY LRMFCIGF+KRR+
Sbjct: 395 LIKSHVDVNTTDNYILRMFCIGFSKRRT 478
Score = 37.4 bits (85), Expect(2) = 8e-13
Identities = 17/27 (62%), Positives = 21/27 (76%)
Frame = +3
Query: 75 GDEDNAFRKIRLRAEDVQGKNLLTNFW 101
GDE++AFR EDVQG+N+LTNFW
Sbjct: 276 GDEEHAFRD-----EDVQGRNVLTNFW 341
>AV773288
Length = 422
Score = 38.1 bits (87), Expect = 0.002
Identities = 23/44 (52%), Positives = 31/44 (70%), Gaps = 1/44 (2%)
Frame = -2
Query: 168 MREIMINQATSCDLKDLVRKFIPE-MIGKEIEKATSSIYPLQNV 210
MR IM + ATSCDLK++VR+F PE G+ + KAT+ +PL V
Sbjct: 421 MRGIMTHPATSCDLKEVVRQFNPEGDWGRGLRKATT--WPLHLV 296
>BP039499
Length = 509
Score = 29.6 bits (65), Expect = 0.55
Identities = 18/60 (30%), Positives = 30/60 (50%), Gaps = 1/60 (1%)
Frame = +2
Query: 24 FAKKDWYDIKAPSVFQVKNVGKTLVSRTQGTKIASEGLK-HRVFEVSLADLQGDEDNAFR 82
F + D+K + Q KN+ + + G K HR+ E++L DLQG E++ F+
Sbjct: 335 FQDIQYCDVK--EIAQKKNLSSLVTVIAFDIETTGFGAKNHRIIEIALRDLQGGENSTFQ 508
>TC11340
Length = 543
Score = 26.9 bits (58), Expect = 3.5
Identities = 19/65 (29%), Positives = 30/65 (45%), Gaps = 3/65 (4%)
Frame = -3
Query: 98 TNFWGMDFTTDKLRSLVRKWQTLIEAHVDVKTTDNYTLRMFCIGFTKRRSNQV---KRTC 154
TN G DF + K + + KW I+A+ V + FCI + + +N + R C
Sbjct: 307 TNLNGSDFFSCK*STPL*KWWQFIQANRMVSGRATQYMLAFCISHSIKCNNTIF*SVRIC 128
Query: 155 YAQSS 159
+ Q S
Sbjct: 127 HLQRS 113
>TC14255
Length = 641
Score = 25.8 bits (55), Expect = 7.9
Identities = 11/29 (37%), Positives = 16/29 (54%)
Frame = -3
Query: 86 LRAEDVQGKNLLTNFWGMDFTTDKLRSLV 114
L E V+ L N WG+ +T +LRS +
Sbjct: 294 LGTESVESNRALVNPWGLQKSTKRLRSFL 208
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.134 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,328,571
Number of Sequences: 28460
Number of extensions: 33653
Number of successful extensions: 164
Number of sequences better than 10.0: 19
Number of HSP's better than 10.0 without gapping: 164
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 164
length of query: 261
length of database: 4,897,600
effective HSP length: 88
effective length of query: 173
effective length of database: 2,393,120
effective search space: 414009760
effective search space used: 414009760
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 54 (25.4 bits)
Medicago: description of AC135798.2


