

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135795.1 - phase: 0
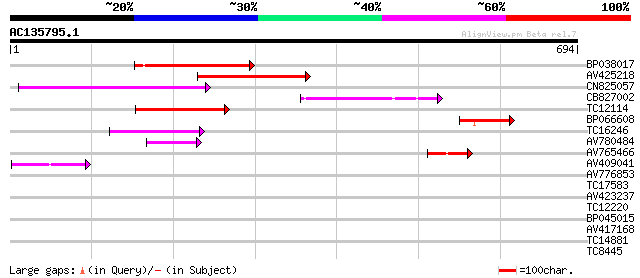
(694 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BP038017 177 5e-45
AV425218 168 2e-42
CN825057 144 4e-35
CB827002 113 9e-26
TC12114 weakly similar to GB|BAC16510.1|23237937|AP005198 transp... 107 6e-24
BP066608 94 1e-19
TC16246 weakly similar to UP|Q8S2M2 (Q8S2M2) Far-red impaired re... 75 3e-14
AV780484 60 2e-09
AV765466 60 2e-09
AV409041 58 4e-09
AV776853 38 0.006
TC17583 weakly similar to UP|Q9LIE5 (Q9LIE5) Far-red impaired re... 37 0.008
AV423237 36 0.024
TC12220 35 0.052
BP045015 30 0.99
AV417168 27 8.4
TC14881 27 8.4
TC8445 similar to UP|O80910 (O80910) At2g38410 protein, partial ... 27 8.4
>BP038017
Length = 570
Score = 177 bits (449), Expect = 5e-45
Identities = 79/147 (53%), Positives = 107/147 (72%)
Frame = +2
Query: 153 YIDKARRLRLGTGDAEAIQNYFVRMQKKNSQFYYVMDVDDKSRLRNVFWVDARCRAAYEY 212
Y+ +++ LG DA+ + NYF +MQ KN FYY + +DD++R+ NVFW DAR R+AY Y
Sbjct: 131 YVKSSQKRTLGR-DAQNLLNYFKKMQGKNPGFYYAIQLDDENRMINVFWADARSRSAYNY 307
Query: 213 FGEVITFDTTYLTNKYDMPFAPFVGVNHHGQSVLLGCALLSNEDTKTFSWLFKTWLECMH 272
FG+ + FDT Y N+Y +PFAPF GVNHHGQ+VL GCALL +E +F+WLF+TWL M+
Sbjct: 308 FGDAVIFDTMYRPNQYQVPFAPFTGVNHHGQNVLFGCALLLDESESSFTWLFRTWLSAMN 487
Query: 273 GRAPNAIITDQDRAMKKAIEDVFPKAR 299
R P +I TDQDRA++ A+ VFP+ R
Sbjct: 488 DRPPVSITTDQDRAIQAAVAQVFPETR 568
>AV425218
Length = 419
Score = 168 bits (426), Expect = 2e-42
Identities = 79/138 (57%), Positives = 99/138 (71%)
Frame = +2
Query: 231 PFAPFVGVNHHGQSVLLGCALLSNEDTKTFSWLFKTWLECMHGRAPNAIITDQDRAMKKA 290
P A VGVNHHGQSVL GCALLS+ED+++F WLF++ L CM G P IITD AMKKA
Sbjct: 5 PLATLVGVNHHGQSVLFGCALLSSEDSESFVWLFQSLLHCMSGVPPQGIITDHSEAMKKA 184
Query: 291 IEDVFPKARHRWCLWHLMKKVPEKLGRHSHYESIKLLLHDAVYDSSSISDFMEKWKKMIE 350
IE V P RHRWCL ++MKK+P+KL ++ YESI+ L + VYD+ I +F WKK++E
Sbjct: 185 IETVLPSTRHRWCLSYIMKKLPQKLLGYAQYESIRHHLQNVVYDAVVIDEFERNWKKIVE 364
Query: 351 CYELHDNEWLKGLFDERY 368
+ L DNEWL LF ER+
Sbjct: 365 DFGLEDNEWLNELFLERH 418
>CN825057
Length = 721
Score = 144 bits (364), Expect = 4e-35
Identities = 76/237 (32%), Positives = 127/237 (53%), Gaps = 2/237 (0%)
Frame = +1
Query: 12 YYKSYARCMGFGTVKINSKNAKDGKKYFT--LGCTRARSYVSNSKNLLKPNPTIRAQCMA 69
+Y+ YA+ MGF T NS+ +K K++ C+R + + + + C A
Sbjct: 10 FYQEYAKSMGFTTSIKNSRRSKKTKEFIDAKFACSRYGVTPESDGGSNRRSSVKKTDCKA 189
Query: 70 RVNMSMSLDGKITITKVALEHNHGLSPTKSRYFRCNKNLDPHTKRRLDINDQAGINVSRN 129
+++ DGK I + EHNH L P + +FR ++N+ K +DI +
Sbjct: 190 CMHVKRKPDGKWIIHEFIKEHNHELLPALAYHFRIHRNVKLAEKNNMDILHAVSERTRKM 369
Query: 130 FRSMVVEANGYDNLTFGEKDCRNYIDKARRLRLGTGDAEAIQNYFVRMQKKNSQFYYVMD 189
+ M ++ G N+ D + K + L + GDA+ + YF +QK+N F+Y +D
Sbjct: 370 YVEMSRQSGGCLNIESLVGDLNDQFKKGQYLAMDEGDAQVMLEYFKHIQKENPNFFYSID 549
Query: 190 VDDKSRLRNVFWVDARCRAAYEYFGEVITFDTTYLTNKYDMPFAPFVGVNHHGQSVL 246
++++ RLRN+FW+DA+ Y F +V++FDT+Y+ + +PFAPFVGVNHH Q +L
Sbjct: 550 LNEEQRLRNIFWIDAKSINDYLSFNDVVSFDTSYIKSNEKLPFAPFVGVNHHCQPIL 720
>CB827002
Length = 534
Score = 113 bits (283), Expect = 9e-26
Identities = 65/174 (37%), Positives = 96/174 (54%)
Frame = +1
Query: 356 DNEWLKGLFDERYRWVPVYVRDTFWAGMSTTQRSESMNSFFDGYVSSKTTLKQFVEQYDN 415
D+EWL L+ + PV++RDTF+A MS TQRS+SMNS+FDGYV++ T L QF + Y+
Sbjct: 31 DHEWLS-LYSSCRQGAPVHLRDTFFAEMSITQRSDSMNSYFDGYVNASTNLNQFFKLYEK 207
Query: 416 ALKDKIEKESMADFVSFNTTIACISLFGFESQFQKAFTNAKFKEFQIEIASMIYCNAFLE 475
AL+ + EKE AD+ + NT + E Q + +T F FQ E+ + F+
Sbjct: 208 ALESRNEKEVRADYDTMNTLPVLRTPSPMEKQASELYTRKIFMRFQEELVGTL---TFMA 378
Query: 476 GMENLNSTFCVIESKKVYDRFKDTRFRVIFNEKDFEIQCACCLFEFKGILCRHI 529
+ + K + K + V FN + + C+C +FEF G+LCRHI
Sbjct: 379 SKADDDGEVITYHVAKFGEDHK--AYYVKFNVLEMKASCSCQMFEFSGLLCRHI 534
>TC12114 weakly similar to GB|BAC16510.1|23237937|AP005198 transposase-like
{Oryza sativa (japonica cultivar-group);}, partial (8%)
Length = 488
Score = 107 bits (267), Expect = 6e-24
Identities = 50/115 (43%), Positives = 76/115 (65%)
Frame = +1
Query: 155 DKARRLRLGTGDAEAIQNYFVRMQKKNSQFYYVMDVDDKSRLRNVFWVDARCRAAYEYFG 214
+K R+ L G+A I YF R +NS FY+ +DD+ ++ NVFWVDAR Y YFG
Sbjct: 10 EKRRQRSLLHGEAGYILQYFQRKLVENSPFYHAYQLDDEDQITNVFWVDARMLIDYGYFG 189
Query: 215 EVITFDTTYLTNKYDMPFAPFVGVNHHGQSVLLGCALLSNEDTKTFSWLFKTWLE 269
++++ D+TY T+ + P A F G NHH ++V+ G ALL +E T+++ WLF+++LE
Sbjct: 190 DMVSLDSTYCTHSSNRPLAVFSGFNHHRKAVIFGAALLYDETTESY*WLFESFLE 354
>BP066608
Length = 530
Score = 93.6 bits (231), Expect = 1e-19
Identities = 47/70 (67%), Positives = 55/70 (78%), Gaps = 3/70 (4%)
Frame = -1
Query: 551 WRKDIKRRHTLIKCGFD---GNVELQLVGKACDAFYEVVSVGIHTEDDLLKVMNRINELK 607
WRKD+KRRHTL +C FD GNVELQ + KACDAF+EVV GI+T++DLLKVMN IN LK
Sbjct: 527 WRKDVKRRHTLTRCXFDHLAGNVELQRLNKACDAFHEVVFAGINTDEDLLKVMNWINXLK 348
Query: 608 IELNCEGSSS 617
IEL +SS
Sbjct: 347 IELTGNEASS 318
>TC16246 weakly similar to UP|Q8S2M2 (Q8S2M2) Far-red impaired response
protein-like, partial (4%)
Length = 660
Score = 75.5 bits (184), Expect = 3e-14
Identities = 39/116 (33%), Positives = 56/116 (47%)
Frame = +2
Query: 123 GINVSRNFRSMVVEANGYDNLTFGEKDCRNYIDKARRLRLGTGDAEAIQNYFVRMQKKNS 182
G+ M+ G+++L F + D N+I K +R R+ GDA A +Y +
Sbjct: 20 GVRACHIMALMLGPKGGHESLGFTKTDLSNHIAKPKRERIQNGDAAAALSYLEGKADNDP 199
Query: 183 QFYYVMDVDDKSRLRNVFWVDARCRAAYEYFGEVITFDTTYLTNKYDMPFAPFVGV 238
F+Y L N+FW D R Y FG+VI FD+TY NKY+ P F+ V
Sbjct: 200 MFFYKFTKTGDESLENLFWCDGVSRMDYNVFGDVIAFDSTYKKNKYNKPLVVFLHV 367
>AV780484
Length = 529
Score = 59.7 bits (143), Expect = 2e-09
Identities = 28/67 (41%), Positives = 39/67 (57%)
Frame = -1
Query: 168 EAIQNYFVRMQKKNSQFYYVMDVDDKSRLRNVFWVDARCRAAYEYFGEVITFDTTYLTNK 227
+A+ YFV Q +N F+Y +D+D L +VFWVD + R YE ++ T YL NK
Sbjct: 205 KAMTEYFVSTQGENPNFFYAIDLDLNRHLTSVFWVDIKGRLDYETSMMLVLIHTHYLKNK 26
Query: 228 YDMPFAP 234
Y +PF P
Sbjct: 25 YKIPFFP 5
>AV765466
Length = 599
Score = 59.7 bits (143), Expect = 2e-09
Identities = 24/55 (43%), Positives = 38/55 (68%)
Frame = -2
Query: 512 IQCACCLFEFKGILCRHILCVLQLTGKTESVPSCYILSRWRKDIKRRHTLIKCGF 566
++C C LFEF+GILCRH L +L + + VP Y+L RWRK+++R++ +K +
Sbjct: 598 VKCDCRLFEFRGILCRHSLAILS-QERVKEVPDRYVLDRWRKNMRRKYVYVKTSY 437
>AV409041
Length = 349
Score = 58.2 bits (139), Expect = 4e-09
Identities = 30/99 (30%), Positives = 50/99 (50%), Gaps = 2/99 (2%)
Frame = +3
Query: 3 FSSEEEVIKYYKSYARCMGFGTVKINSKNAKDGKKYF--TLGCTRARSYVSNSKNLLKPN 60
F S E +YK YA+ GFGT K++S+ ++ K++ C R + S + + P
Sbjct: 51 FESHEAAYAFYKEYAKSAGFGTAKLSSRRSRASKEFIDAKFSCIRYGN-KQQSDDAINPR 227
Query: 61 PTIRAQCMARVNMSMSLDGKITITKVALEHNHGLSPTKS 99
P+ + C A +++ DGK + EHNH L P ++
Sbjct: 228 PSPKIGCKASMHVKRRQDGKWYVYSFVKEHNHELLPAQA 344
>AV776853
Length = 625
Score = 37.7 bits (86), Expect = 0.006
Identities = 29/115 (25%), Positives = 48/115 (41%), Gaps = 13/115 (11%)
Frame = +1
Query: 1 MTFSSEEEVIKYYKSYARCMGFGTVKINSKNAKDGKKYFTLGCTRARSYVSNSKNLLKP- 59
+ F SEEE ++Y++YA+ GF K + G+ Y G R +V N + L
Sbjct: 238 LDFGSEEEAYQFYQAYAKYQGFIVRKDDI-----GRDYH--GNVNMRQFVCNRQGLRSKK 396
Query: 60 -----------NPTIRAQCMARVNMSMSLD-GKITITKVALEHNHGLSPTKSRYF 102
P C+A++ + + GK + HNH L+P + +F
Sbjct: 397 HYNRTDRKRDHKPVTHTNCLAKLRVHLDYKIGKWKVVSFEECHNHELTPARFVHF 561
>TC17583 weakly similar to UP|Q9LIE5 (Q9LIE5) Far-red impaired response
protein, mutator-like transposase-like protein,
phytochrome A signaling protein-like, partial (3%)
Length = 666
Score = 37.4 bits (85), Expect = 0.008
Identities = 22/61 (36%), Positives = 30/61 (49%), Gaps = 5/61 (8%)
Frame = +3
Query: 210 YEYFGEVITFDTTYLTNKYDMPFAP-----FVGVNHHGQSVLLGCALLSNEDTKTFSWLF 264
YEYFG+V++ DTTY TN P A + +N +LL C + + SW F
Sbjct: 18 YEYFGDVVSLDTTYSTNNAYRPLAVSMCF*AICLNFSILLLLLYCVCIGKHLPQ*QSWCF 197
Query: 265 K 265
K
Sbjct: 198 K 200
>AV423237
Length = 310
Score = 35.8 bits (81), Expect = 0.024
Identities = 28/87 (32%), Positives = 43/87 (49%), Gaps = 1/87 (1%)
Frame = +2
Query: 203 DARCRAAYEYFGEVITFDTTYLTNKYDMPFAPFVGVNHHGQSVLLGCALLSNEDTKTFSW 262
DA R Y YFG+ + FDTT + N + + + G + V+ AL++NE +F W
Sbjct: 17 DATSRMNYSYFGDAVIFDTT-IDNHIESICSSW-G*SSWATCVIWLVALIANESESSFVW 190
Query: 263 LFKTW-LECMHGRAPNAIITDQDRAMK 288
L + C+ A +I TD D + K
Sbjct: 191 LSGLGSMPCLD--ATCSITTDLDHSYK 265
>TC12220
Length = 473
Score = 34.7 bits (78), Expect = 0.052
Identities = 16/35 (45%), Positives = 23/35 (65%)
Frame = +1
Query: 520 EFKGILCRHILCVLQLTGKTESVPSCYILSRWRKD 554
E++G+LCRH L V + + VPS YIL RW ++
Sbjct: 1 EYEGVLCRHPLRVFXIL-ELREVPSRYILHRWTRN 102
>BP045015
Length = 527
Score = 30.4 bits (67), Expect = 0.99
Identities = 15/35 (42%), Positives = 22/35 (62%)
Frame = -1
Query: 240 HHGQSVLLGCALLSNEDTKTFSWLFKTWLECMHGR 274
HH + +LLG + S + TFSWL+ L+C+H R
Sbjct: 266 HHKEIILLGLSRKSLQLEGTFSWLYH--LKCIHFR 168
>AV417168
Length = 395
Score = 27.3 bits (59), Expect = 8.4
Identities = 19/61 (31%), Positives = 26/61 (42%), Gaps = 2/61 (3%)
Frame = +1
Query: 34 DGKKYFTLGCTR--ARSYVSNSKNLLKPNPTIRAQCMARVNMSMSLDGKITITKVALEHN 91
DG K TL RS SK L P P + ++ + S S + K T+T HN
Sbjct: 193 DGSKQATLAARTP*GRSRTIPSKELHFPPPQPAEKTFSKAHSSPSQETKTTVTTTTNNHN 372
Query: 92 H 92
+
Sbjct: 373 N 375
>TC14881
Length = 609
Score = 27.3 bits (59), Expect = 8.4
Identities = 10/15 (66%), Positives = 10/15 (66%)
Frame = +3
Query: 514 CACCLFEFKGILCRH 528
C C FKGILCRH
Sbjct: 444 CVCLRVMFKGILCRH 488
>TC8445 similar to UP|O80910 (O80910) At2g38410 protein, partial (7%)
Length = 841
Score = 27.3 bits (59), Expect = 8.4
Identities = 13/36 (36%), Positives = 18/36 (49%)
Frame = +2
Query: 91 NHGLSPTKSRYFRCNKNLDPHTKRRLDINDQAGINV 126
NH LSP + + N NL +T L+ N IN+
Sbjct: 152 NHNLSPNPNTFSNLNHNLSSNTFSNLNHNRNFRINI 259
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.322 0.135 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,035,245
Number of Sequences: 28460
Number of extensions: 167216
Number of successful extensions: 893
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 879
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 887
length of query: 694
length of database: 4,897,600
effective HSP length: 97
effective length of query: 597
effective length of database: 2,136,980
effective search space: 1275777060
effective search space used: 1275777060
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC135795.1


