

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135565.15 + phase: 0
(557 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
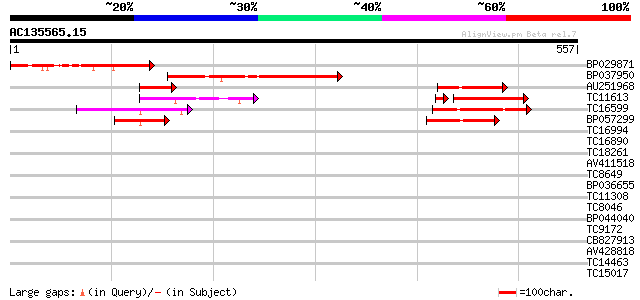
Score E
Sequences producing significant alignments: (bits) Value
BP029871 201 2e-52
BP037950 189 1e-48
AU251968 119 1e-27
TC11613 similar to UP|Q8W240 (Q8W240) GT-2 factor (Fragment), pa... 99 7e-22
TC16599 similar to UP|Q8W239 (Q8W239) GT-2 factor (Fragment), pa... 77 7e-15
BP057299 69 3e-12
TC16994 similar to UP|O22805 (O22805) At2g33550 protein, partial... 40 0.001
TC16890 38 0.005
TC18261 similar to UP|WSC2_YEAST (P53832) Cell wall integrity an... 37 0.006
AV411518 37 0.006
TC8649 similar to PIR|T52450|T52450 ribosomal protein S9 [import... 36 0.014
BP036655 36 0.019
TC11308 similar to UP|CENB_HUMAN (P07199) Major centromere autoa... 36 0.019
TC8046 similar to UP|Q7XJC2 (Q7XJC2) Adenine phosphoribosyltrans... 36 0.019
BP044040 35 0.024
TC9172 35 0.024
CB827913 34 0.054
AV428818 34 0.071
TC14463 similar to UP|Q9SLF1 (Q9SLF1) Nodulin-like protein (At2g... 33 0.093
TC15017 33 0.12
>BP029871
Length = 519
Score = 201 bits (512), Expect = 2e-52
Identities = 113/154 (73%), Positives = 122/154 (78%), Gaps = 12/154 (7%)
Frame = +3
Query: 1 MYDGVPDQFHQFITPRTSSSSLPLHLPFPL---STPNN--TFPPFDPYNQQNHPSQHHQL 55
M+DGVPDQFHQFITPRTS LPLHLPFPL S PNN TFP FDPYN PS HHQ
Sbjct: 90 MFDGVPDQFHQFITPRTS---LPLHLPFPLHASSAPNNNTTFPSFDPYN----PS-HHQX 245
Query: 56 PLQVQPNLLH-PLHPHKDDEDKEQNST--PSMNNFQIDRDQRQILPQL----IDPWTNDE 108
P Q PNLLH PLH HKD+E +E N+T P+ NFQI+RDQRQ LPQL +DPWTNDE
Sbjct: 246 PFQ--PNLLHHPLH-HKDEEKEENNNTVQPNPMNFQIERDQRQQLPQLQQQQVDPWTNDE 416
Query: 109 VLALLKIRSSMESWFPDFTWEHVSRKLAEVGYKR 142
VLALL+IRSSMESWFP+FTWEHVSRKLAEVGYKR
Sbjct: 417 VLALLRIRSSMESWFPEFTWEHVSRKLAEVGYKR 518
Score = 33.5 bits (75), Expect = 0.093
Identities = 33/123 (26%), Positives = 46/123 (36%), Gaps = 19/123 (15%)
Frame = +3
Query: 361 SQNPNPETLKKTLQPIPENPSSTLPSSSTTLVAQP---------RNNNPISSYSLISSGE 411
S PN T + P NPS L+ P NNN + + E
Sbjct: 177 SSAPNNNTTFPSFDPY--NPSHHQXPFQPNLLHHPLHHKDEEKEENNNTVQPNPMNFQIE 350
Query: 412 RDDIGRR----------WPKDEVLALINLRCNNNNEEKEGNSNNKAPLWERISQGMLELG 461
RD + W DEVLAL+ +R + + E WE +S+ + E+G
Sbjct: 351 RDQRQQLPQLQQQQVDPWTNDEVLALLRIRSSMESWFPEFT-------WEHVSRKLAEVG 509
Query: 462 YKR 464
YKR
Sbjct: 510 YKR 518
>BP037950
Length = 529
Score = 189 bits (479), Expect = 1e-48
Identities = 105/180 (58%), Positives = 125/180 (69%), Gaps = 8/180 (4%)
Frame = +1
Query: 156 RFFNNINHNQNSFG-KNFRFVTELEEVYQGGGGENNKNLVEAEKQNEVQDKM-------D 207
R+FNNIN+ S N+RF+TELEE+Y+ G E + ++Q + QDKM D
Sbjct: 1 RYFNNINYTNKSTNINNYRFLTELEELYEEVGTEKSTT---HQQQQQQQDKMEERAALDD 171
Query: 208 PHEEDSRMDDVLVSKKSEEEVVEKGTTNDEKKRKRSGDDRFEVFKGFCESVVKKMMDQQE 267
D +D + EE EK E+KRKR DDRFEVFKGFCESVV MM +QE
Sbjct: 172 QDHHDDSVDQTKQNNAQSEEDQEK--IMKERKRKRV-DDRFEVFKGFCESVVNNMMGRQE 342
Query: 268 EMHNKLIEDMVKRDEEKFSREEAWKKQEMEKMNKELELMAHEQAIAGDRQAHIIQFLNKF 327
EMH K+IEDMV+RDEEKFS+EEAWKKQEM+KMNKELE+MAHEQAIAGDRQA+IIQFL F
Sbjct: 343 EMHRKIIEDMVRRDEEKFSKEEAWKKQEMDKMNKELEMMAHEQAIAGDRQANIIQFLKNF 522
>AU251968
Length = 261
Score = 119 bits (299), Expect = 1e-27
Identities = 60/70 (85%), Positives = 63/70 (89%), Gaps = 1/70 (1%)
Frame = +3
Query: 421 KDEVLALINLRC-NNNNEEKEGNSNNKAPLWERISQGMLELGYKRSAKRCKEKWENINKY 479
K+EV ALINLRC +NNNE+KEGN NKAPLWERISQGMLELGYKRSAKRCKEKWENINKY
Sbjct: 57 KNEVWALINLRCTSNNNEDKEGN--NKAPLWERISQGMLELGYKRSAKRCKEKWENINKY 230
Query: 480 FRKTKDANRK 489
FRKTKD K
Sbjct: 231 FRKTKDNTSK 260
Score = 45.4 bits (106), Expect = 2e-05
Identities = 17/37 (45%), Positives = 27/37 (72%)
Frame = +3
Query: 128 WEHVSRKLAEVGYKRSAEKCKEKFEEESRFFNNINHN 164
WE +S+ + E+GYKRSA++CKEK+E +++F N
Sbjct: 141 WERISQGMLELGYKRSAKRCKEKWENINKYFRKTKDN 251
>TC11613 similar to UP|Q8W240 (Q8W240) GT-2 factor (Fragment), partial (26%)
Length = 731
Score = 98.6 bits (244), Expect(2) = 7e-22
Identities = 43/73 (58%), Positives = 53/73 (71%)
Frame = +3
Query: 437 EEKEGNSNNKAPLWERISQGMLELGYKRSAKRCKEKWENINKYFRKTKDANRKRSLDSRT 496
E K + KAPLWE IS M +GY R+AKRC EKWENINKYF+K K++N++R DS+T
Sbjct: 108 EAKYQENGPKAPLWEEISAAMQRIGYNRNAKRCNEKWENINKYFKKVKESNKQRREDSKT 287
Query: 497 CPYFHLLTNLYNQ 509
CPYFH L LY +
Sbjct: 288 CPYFHELEALYKE 326
Score = 45.4 bits (106), Expect = 2e-05
Identities = 32/128 (25%), Positives = 58/128 (45%), Gaps = 11/128 (8%)
Frame = +3
Query: 128 WEHVSRKLAEVGYKRSAEKCKEKFEEESRFFNNI---NHNQNSFGKNFRFVTELEEVYQG 184
WE +S + +GY R+A++C EK+E +++F + N + K + ELE +Y+
Sbjct: 147 WEEISAAMQRIGYNRNAKRCNEKWENINKYFKKVKESNKQRREDSKTCPYFHELEALYK- 323
Query: 185 GGGENNKNLVEAEKQNEVQDKMDPHEEDSRMDDVLVSKKS--------EEEVVEKGTTND 236
+NL A N ++M M+ ++V + EE V + TN
Sbjct: 324 -EKSKTQNLFGASIHNMRTNEM--------MEPLMVQPEQQWRPPPQYEESVKKNDETNA 476
Query: 237 EKKRKRSG 244
++ +R G
Sbjct: 477 REQDRRRG 500
Score = 22.3 bits (46), Expect(2) = 7e-22
Identities = 9/13 (69%), Positives = 10/13 (76%)
Frame = +2
Query: 419 WPKDEVLALINLR 431
WPK EV ALI +R
Sbjct: 59 WPKAEVHALIKIR 97
>TC16599 similar to UP|Q8W239 (Q8W239) GT-2 factor (Fragment), partial (37%)
Length = 693
Score = 77.0 bits (188), Expect = 7e-15
Identities = 40/97 (41%), Positives = 61/97 (62%)
Frame = +1
Query: 416 GRRWPKDEVLALINLRCNNNNEEKEGNSNNKAPLWERISQGMLELGYKRSAKRCKEKWEN 475
G RWP+ E LAL+ +R + + ++ S+ K PLWE +S+ + LGY RSAK+CKEK+EN
Sbjct: 193 GSRWPRQETLALLKIRSDMDAVFRD--SSLKGPLWEEVSRKLAGLGYHRSAKKCKEKFEN 366
Query: 476 INKYFRKTKDANRKRSLDSRTCPYFHLLTNLYNQGKL 512
+ KY ++TK+ +R + +T +F L L Q L
Sbjct: 367 VYKYHKRTKE-SRSAKPEGKTYRFFDQLQALEKQFSL 474
Score = 74.3 bits (181), Expect = 5e-14
Identities = 44/121 (36%), Positives = 62/121 (50%), Gaps = 7/121 (5%)
Frame = +1
Query: 66 PLHPHKDDEDKEQNSTPSMNNFQIDRDQRQILPQLIDPWTNDEVLALLKIRSSMESWFPD 125
PL P E+ ++D D + W E LALLKIRS M++ F D
Sbjct: 88 PLPPQDGANGGEEKGREGSGGGELDEDGGDKIGSGGSRWPRQETLALLKIRSDMDAVFRD 267
Query: 126 FT-----WEHVSRKLAEVGYKRSAEKCKEKFEEESRFFNNINHNQNS--FGKNFRFVTEL 178
+ WE VSRKLA +GY RSA+KCKEKFE ++ ++++ GK +RF +L
Sbjct: 268 SSLKGPLWEEVSRKLAGLGYHRSAKKCKEKFENVYKYHKRTKESRSAKPEGKTYRFFDQL 447
Query: 179 E 179
+
Sbjct: 448 Q 450
>BP057299
Length = 551
Score = 68.6 bits (166), Expect = 3e-12
Identities = 30/72 (41%), Positives = 49/72 (67%)
Frame = +3
Query: 410 GERDDIGRRWPKDEVLALINLRCNNNNEEKEGNSNNKAPLWERISQGMLELGYKRSAKRC 469
G+R G RWP+ E LAL+ +R + + ++ + K PLW+ +S+ + +LGY R+AK+C
Sbjct: 342 GDRSFGGNRWPRQETLALLKIRSDMDVAFRDASV--KGPLWDEVSRKLADLGYHRNAKKC 515
Query: 470 KEKWENINKYFR 481
KEK+EN+ KY +
Sbjct: 516 KEKFENVYKYHK 551
Score = 62.4 bits (150), Expect = 2e-10
Identities = 31/59 (52%), Positives = 40/59 (67%), Gaps = 5/59 (8%)
Frame = +3
Query: 104 WTNDEVLALLKIRSSMESWFPDFT-----WEHVSRKLAEVGYKRSAEKCKEKFEEESRF 157
W E LALLKIRS M+ F D + W+ VSRKLA++GY R+A+KCKEKFE ++
Sbjct: 369 WPRQETLALLKIRSDMDVAFRDASVKGPLWDEVSRKLADLGYHRNAKKCKEKFENVYKY 545
>TC16994 similar to UP|O22805 (O22805) At2g33550 protein, partial (36%)
Length = 680
Score = 40.0 bits (92), Expect = 0.001
Identities = 24/100 (24%), Positives = 47/100 (47%), Gaps = 5/100 (5%)
Frame = +2
Query: 418 RWPKDEVLALINLRCNNNNEEKEGNSNNKA-----PLWERISQGMLELGYKRSAKRCKEK 472
RW + E+L LI + + + K G + A P W +S + G R +C+++
Sbjct: 254 RWTRQEILVLIQGKADAESRFKPGRAAGSAFGSSEPKWALVSSYCKKHGVNREPVQCRKR 433
Query: 473 WENINKYFRKTKDANRKRSLDSRTCPYFHLLTNLYNQGKL 512
W N+ ++K K+ + S+ T ++ + +L + KL
Sbjct: 434 WSNLAGDYKKIKE--WESSVKDETESFWLMRNDLRRERKL 547
>TC16890
Length = 965
Score = 37.7 bits (86), Expect = 0.005
Identities = 38/164 (23%), Positives = 73/164 (44%)
Frame = +2
Query: 145 EKCKEKFEEESRFFNNINHNQNSFGKNFRFVTELEEVYQGGGGENNKNLVEAEKQNEVQD 204
EK KEK +E N +NS K F + +++ GG +NN+ +V+
Sbjct: 65 EKEKEKGKEREE-----NKEENSKKKGF*LLNKMQG---GGEQQNNQLVVQNSGSLSFSS 220
Query: 205 KMDPHEEDSRMDDVLVSKKSEEEVVEKGTTNDEKKRKRSGDDRFEVFKGFCESVVKKMMD 264
+M +E+ + K EEE+ +++K ++ ++ G E K++
Sbjct: 221 QMSKEDEEMSRSALSNFKAKEEEI---------ERKKMEVREKVQLQLGRVEEETKRLAT 373
Query: 265 QQEEMHNKLIEDMVKRDEEKFSREEAWKKQEMEKMNKELELMAH 308
+EE+ + L + M K E A ++ ++ +NKEL+ + H
Sbjct: 374 IREELES-LADPMRK--------EVAIVRKRIDSVNKELKPLGH 478
>TC18261 similar to UP|WSC2_YEAST (P53832) Cell wall integrity and stress
response component 2 precursor, partial (5%)
Length = 538
Score = 37.4 bits (85), Expect = 0.006
Identities = 23/77 (29%), Positives = 42/77 (53%)
Frame = +3
Query: 334 SSLTSMSTQLQAYLATLTSNSSSSTLHSQNPNPETLKKTLQPIPENPSSTLPSSSTTLVA 393
+SL+ M++ ++ S SS+++ S +P+P ++K T+ + +S+ P SS +
Sbjct: 237 TSLSLMTSTTSTSSSSPNSRSSAASASSSSPSPSSVKTTVPGSTASNTSSRPVSSKSTAI 416
Query: 394 QPRNNNPISSYSLISSG 410
PR ISS + SSG
Sbjct: 417 *PRGRPTISSATSPSSG 467
>AV411518
Length = 360
Score = 37.4 bits (85), Expect = 0.006
Identities = 24/70 (34%), Positives = 32/70 (45%)
Frame = +3
Query: 334 SSLTSMSTQLQAYLATLTSNSSSSTLHSQNPNPETLKKTLQPIPENPSSTLPSSSTTLVA 393
S+L S ++ + S S SST S PNP + P P PS P+SS+ A
Sbjct: 132 SALASWASPCAPTSSGPASPSPSSTAPSPRPNPSSTSGPTSPTPHTPSPPPPTSSSPSSA 311
Query: 394 QPRNNNPISS 403
P + P SS
Sbjct: 312 IPPTSAPSSS 341
>TC8649 similar to PIR|T52450|T52450 ribosomal protein S9 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(81%)
Length = 939
Score = 36.2 bits (82), Expect = 0.014
Identities = 38/142 (26%), Positives = 63/142 (43%), Gaps = 14/142 (9%)
Frame = +2
Query: 338 SMSTQLQAYLATLTSNSSSSTLHSQNPNPETLKKTLQP-----IPENPSSTLPSS----- 387
+M+ L ATL+S S SS + S NP T +T QP +P+ PS T+ ++
Sbjct: 41 TMAVSLSTLTATLSSLSFSSNI-SHNPQTLTYARTTQPLSLSRVPKTPSLTVSAAAPVEP 217
Query: 388 -STTLVAQPRNNNP--ISSYSLISSGERDDIGRRWPKDEVLALINLRCNNNNEEKEGNSN 444
+ L+ ++ P ++ + I +G R R E + + E +GN
Sbjct: 218 ETENLMKYVKSRLPGGFAAQTTIGTGRRKSAIARVVLQEGTGKFIINYRDAKEYLQGN-- 391
Query: 445 NKAPLW-ERISQGMLELGYKRS 465
PLW + I ++ LGY+ S
Sbjct: 392 ---PLWLQYIKVPLVTLGYETS 448
>BP036655
Length = 556
Score = 35.8 bits (81), Expect = 0.019
Identities = 27/77 (35%), Positives = 39/77 (50%)
Frame = +1
Query: 327 FSTSANSSSLTSMSTQLQAYLATLTSNSSSSTLHSQNPNPETLKKTLQPIPENPSSTLPS 386
FST +SSS +S S + L++ TS+S SS+L S + + S+T S
Sbjct: 220 FSTPHSSSSSSSFSVSISVSLSSSTSSSLSSSLISSS-------------SLHSSTTSSS 360
Query: 387 SSTTLVAQPRNNNPISS 403
SSTT + P + P SS
Sbjct: 361 SSTTSSSSPSSERPSSS 411
Score = 27.7 bits (60), Expect = 5.1
Identities = 20/60 (33%), Positives = 32/60 (53%)
Frame = +1
Query: 308 HEQAIAGDRQAHIIQFLNKFSTSANSSSLTSMSTQLQAYLATLTSNSSSSTLHSQNPNPE 367
H + + I L+ ++S+ SSSL S S+ L + T++SSSST S +P+ E
Sbjct: 232 HSSSSSSSFSVSISVSLSSSTSSSLSSSLISSSS-----LHSSTTSSSSSTTSSSSPSSE 396
>TC11308 similar to UP|CENB_HUMAN (P07199) Major centromere autoantigen B
(Centromere protein B) (CENP-B), partial (4%)
Length = 756
Score = 35.8 bits (81), Expect = 0.019
Identities = 24/51 (47%), Positives = 27/51 (52%), Gaps = 1/51 (1%)
Frame = +2
Query: 22 LPLHLPFPLSTPNNTFPPFDP-YNQQNHPSQHHQLPLQVQPNLLHPLHPHK 71
LPLHLP PL P PP P Y+ Q+H H L LQV P PL P +
Sbjct: 254 LPLHLPRPLPLP----PPLPPHYHFQSHGICFH-LHLQVPPGHFLPLDPQR 391
>TC8046 similar to UP|Q7XJC2 (Q7XJC2) Adenine phosphoribosyltransferase
(Fragment), partial (85%)
Length = 1298
Score = 35.8 bits (81), Expect = 0.019
Identities = 22/73 (30%), Positives = 39/73 (53%), Gaps = 1/73 (1%)
Frame = +1
Query: 325 NKFSTSANSSSLTSMSTQLQAYLATLTSNSSSSTLHSQNPNPETLKKTLQPIPENPS-ST 383
N S+S++SSS + +T +S+SSS+ + SQ+P T+ ++ IP+ P
Sbjct: 178 NSTSSSSSSSSSSIRFQNSPPLRSTASSSSSSANMASQDPRIATISSAIRVIPDFPKPGI 357
Query: 384 LPSSSTTLVAQPR 396
+ TTL+ P+
Sbjct: 358 MFQDITTLLLDPK 396
>BP044040
Length = 491
Score = 35.4 bits (80), Expect = 0.024
Identities = 33/112 (29%), Positives = 53/112 (46%), Gaps = 7/112 (6%)
Frame = +3
Query: 312 IAGDRQAHIIQFLNKFSTSANSSSLTSMSTQLQAYLATLTSNSSSSTLH-SQNPNPETLK 370
I GD + + F+ K + SSS Q + ++ +S+SSS+ ++ ++NP P L
Sbjct: 48 IKGDMKDKVKGFMKKVNNPFTSSSSGKFKGQGRVLGSSSSSSSSSAPINPNRNPTPR-LT 224
Query: 371 KTLQPIPENPSSTLPSSSTTLVAQPR--NNNPISSY----SLISSGERDDIG 416
T P P S P+S+T P+ N NP + SL++S R G
Sbjct: 225 PTQNPNPN--SKPRPASTTNHEPSPQKTNRNPGDGFDPFDSLVTSSNRSQNG 374
>TC9172
Length = 605
Score = 35.4 bits (80), Expect = 0.024
Identities = 16/47 (34%), Positives = 25/47 (53%)
Frame = +3
Query: 449 LWERISQGMLELGYKRSAKRCKEKWENINKYFRKTKDANRKRSLDSR 495
LWE ISQ +L G RS +CK W ++ + + K+ + +S R
Sbjct: 39 LWEEISQNLLSDGISRSPGQCKSLWTSLVLKYEEIKNTDGNKSWQYR 179
>CB827913
Length = 567
Score = 34.3 bits (77), Expect = 0.054
Identities = 29/85 (34%), Positives = 39/85 (45%), Gaps = 6/85 (7%)
Frame = -3
Query: 15 PRTSSSSLPLHLPFPLS-----TPNNTFPPFDPYNQQNHP-SQHHQLPLQVQPNLLHPLH 68
P+ SS LH F LS +P F F+P+ +HP QH QL + NL H LH
Sbjct: 271 PQEESS---LHFLFQLSQLQH*SPFQKFQLFEPFPFHSHPLLQHQQL---LSGNLSHQLH 110
Query: 69 PHKDDEDKEQNSTPSMNNFQIDRDQ 93
P + + + P + FQ DQ
Sbjct: 109 PRQPHWPR-PSQMPQLPPFQHQEDQ 38
>AV428818
Length = 417
Score = 33.9 bits (76), Expect = 0.071
Identities = 26/77 (33%), Positives = 36/77 (45%), Gaps = 1/77 (1%)
Frame = +3
Query: 328 STSANSS-SLTSMSTQLQAYLATLTSNSSSSTLHSQNPNPETLKKTLQPIPENPSSTLPS 386
STS+N + + ST L+ TS S++S S +PN +T T P N S T P
Sbjct: 150 STSSNPKPTRNASSTSAAPPLSPRTSTSTASPSPSPSPNSKTA--TTSPPSTNSSGTSPP 323
Query: 387 SSTTLVAQPRNNNPISS 403
S+ T + NP S
Sbjct: 324 SALTSATSVSSRNPSCS 374
>TC14463 similar to UP|Q9SLF1 (Q9SLF1) Nodulin-like protein
(At2g16660/T24I21.7), partial (40%)
Length = 1122
Score = 33.5 bits (75), Expect = 0.093
Identities = 18/40 (45%), Positives = 23/40 (57%), Gaps = 1/40 (2%)
Frame = +2
Query: 352 SNSSSSTLHSQNPNPETLKKT-LQPIPENPSSTLPSSSTT 390
+ S+SS NP+P T + P P NPS T PSSS+T
Sbjct: 146 NGSASSPPSGSNPSPATTTPSPTTPTPSNPSCTSPSSSST 265
>TC15017
Length = 1155
Score = 33.1 bits (74), Expect = 0.12
Identities = 18/46 (39%), Positives = 22/46 (47%)
Frame = +1
Query: 24 LHLPFPLSTPNNTFPPFDPYNQQNHPSQHHQLPLQVQPNLLHPLHP 69
LH P LS N PP P + PS HQLP + ++ P HP
Sbjct: 97 LHTPSHLSLRNQNHPP--PPRLLSFPSSQHQLPRNLSNGVV*PPHP 228
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.309 0.127 0.365
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,683,636
Number of Sequences: 28460
Number of extensions: 152647
Number of successful extensions: 1703
Number of sequences better than 10.0: 148
Number of HSP's better than 10.0 without gapping: 1491
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1618
length of query: 557
length of database: 4,897,600
effective HSP length: 95
effective length of query: 462
effective length of database: 2,193,900
effective search space: 1013581800
effective search space used: 1013581800
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC135565.15


