

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135505.8 + phase: 0
(454 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
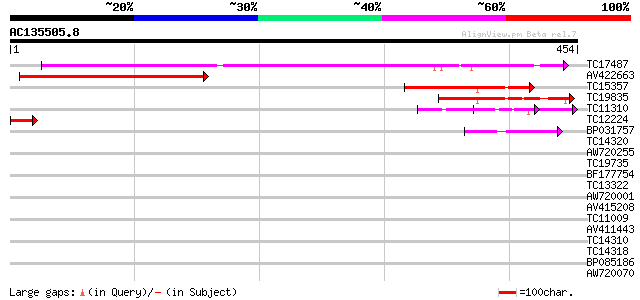
Score E
Sequences producing significant alignments: (bits) Value
TC17487 UP|CAE45597 (CAE45597) SAR DNA-binding protein-like prot... 323 5e-89
AV422663 263 4e-71
TC15357 similar to UP|Q9ZV06 (Q9ZV06) At2g39020 protein, partial... 150 4e-37
TC19835 similar to UP|Q9ZRW0 (Q9ZRW0) Nucleolar protein (Fragmen... 122 1e-28
TC11310 similar to UP|BAD11331 (BAD11331) BRI1-KD interacting pr... 51 5e-07
TC12224 similar to PIR|D96602|D96602 nucleolar protein [imported... 44 4e-05
BP031757 40 8e-04
TC14320 similar to UP|Q41111 (Q41111) Dehydrin, partial (56%) 39 0.002
AW720255 38 0.003
TC19735 similar to UP|Q8NJM5 (Q8NJM5) Probable ATP-dependent RNA... 38 0.003
BF177754 37 0.007
TC13322 similar to UP|Q9ZS13 (Q9ZS13) RNA helicase (Fragment), p... 37 0.009
AW720001 34 0.057
AV415208 33 0.13
TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Ar... 33 0.13
AV411443 32 0.17
TC14310 homologue to UP|Q9M3H6 (Q9M3H6) Histone H2B, partial (92%) 32 0.17
TC14318 similar to UP|O64739 (O64739) Ran-binding protein (AtRan... 32 0.22
BP085186 32 0.28
AW720070 32 0.28
>TC17487 UP|CAE45597 (CAE45597) SAR DNA-binding protein-like protein,
complete
Length = 1794
Score = 323 bits (827), Expect = 5e-89
Identities = 189/453 (41%), Positives = 266/453 (57%), Gaps = 31/453 (6%)
Frame = +1
Query: 26 LGVAESKIGSHIHEATKIPVQSNEFVGELIRGVRQHFDKFVGDLKQGDLEKAQLGLCHSY 85
L VA+SK+G+ I E KI N V EL+RGVR ++ + L D+ LGL HS
Sbjct: 391 LAVADSKLGNMIKEKLKIDCVHNNAVMELMRGVRNQLNELISGLAVQDMAPMSLGLSHSL 570
Query: 86 SRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFAMRVREWYSWHFPELVKIVNDNYLYCK 145
SR K+KF+ +VD MV+QAI LLD LDK++N++AMRVREWY WHFPEL KI+ DN Y K
Sbjct: 571 SRYKLKFSAEKVDTMVVQAIGLLDDLDKELNTYAMRVREWYGWHFPELTKIIQDNIQYAK 750
Query: 146 VAKFIEDKSKLAEDKIESLTDLVGDEDKAKEIVEAAKASMGQDLSPVDLINVHMFAQRVM 205
K + D+ A+ + E+ +E+ EA+ SMG ++ +DL+N+ +V+
Sbjct: 751 AVKLMGDRVNAAQTDFSEVLP----EEVEEEVKEASVISMGTEIGELDLMNIKELCDQVL 918
Query: 206 DLSDYRRRLSDYLSTKMNDIAPNLQSLVGDSVGARLISHAGSLTNLAKCPSSTLQILGAE 265
LS+YR +L DYL ++MN IAPNL +LVG+ VGARLI+H GSL NLAK P ST+QILGAE
Sbjct: 919 SLSEYRAQLYDYLKSRMNSIAPNLTALVGELVGARLIAHGGSLINLAKQPGSTVQILGAE 1098
Query: 266 KALFRALKTRGNTPKYGLIFHSSFIGRASARNKGRMARYLANKCSIASRIDCFSEKGTSA 325
KALFRALKT+ TPKYGLI+H+S IG+A+ + KG+++R LA K ++A R D + +
Sbjct: 1099KALFRALKTKHATPKYGLIYHASLIGQAAPKLKGKISRSLAAKAALAIRCDALGDGVDNT 1278
Query: 326 FGEKLREQVEERL---------------------DFYDKG--------VAPRKNIDVMKS 356
G + R ++E RL + YDK + P K +
Sbjct: 1279MGLENRAKLELRLRNLEGKELGRFAGSAKGKPKIEAYDKDRKKGAGGLITPAKTYNPSAD 1458
Query: 357 AIESVDNKDTEM--ETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKK 414
A+ S D+ M +T E SA K K+KK+K + + + D ++ K KK+
Sbjct: 1459AVIS-QKSDSAMDEDTHEPSADKKKEKKEKKKKKEKNEEKDVPLLADEDGDEEKEVVKKE 1635
Query: 415 KKEKRKLDQEVEVEDKVVEDGANADSSKKKKKK 447
KK+KRK E+ +++G +A KKK+KK
Sbjct: 1636KKKKRK----ESTENVELQNGDDAGEKKKKRKK 1722
>AV422663
Length = 457
Score = 263 bits (672), Expect = 4e-71
Identities = 130/151 (86%), Positives = 138/151 (91%)
Frame = +3
Query: 9 LETNLPKVKEGKKAKFSLGVAESKIGSHIHEATKIPVQSNEFVGELIRGVRQHFDKFVGD 68
LE NLPKVKEGKK KFSLGV++ KIGS I E +KIP QSNEFV EL+RGVR HF+KF+GD
Sbjct: 3 LEKNLPKVKEGKKPKFSLGVSDPKIGSQIAEVSKIPCQSNEFVSELLRGVRLHFNKFIGD 182
Query: 69 LKQGDLEKAQLGLCHSYSRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFAMRVREWYSW 128
LK GDLEKAQLGL HSYSRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSF+MRVREWYSW
Sbjct: 183 LKTGDLEKAQLGLGHSYSRAKVKFNVNRVDNMVIQAIFLLDTLDKDVNSFSMRVREWYSW 362
Query: 129 HFPELVKIVNDNYLYCKVAKFIEDKSKLAED 159
HFPELVKIVNDNYLY KVAKFI+DKSKL ED
Sbjct: 363 HFPELVKIVNDNYLYAKVAKFIKDKSKLTED 455
>TC15357 similar to UP|Q9ZV06 (Q9ZV06) At2g39020 protein, partial (69%)
Length = 1578
Score = 150 bits (379), Expect = 4e-37
Identities = 80/109 (73%), Positives = 92/109 (84%), Gaps = 5/109 (4%)
Frame = +2
Query: 317 CFSEKGTSAFGEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEV--- 373
CFSE ++AFG KLREQVEERLDFYDKGVAPRKNIDVMKSAIES N D EMETEEV
Sbjct: 2 CFSESSSTAFGLKLREQVEERLDFYDKGVAPRKNIDVMKSAIESAANNDAEMETEEVPVE 181
Query: 374 -SAKKTKKKKQKAADGDDMAVDKAAEITNGDA-EDHKSEKKKKKKEKRK 420
S+KK+KKK++ A +GD+MAVDK +TNGDA EDHKSEKKKKKK+K++
Sbjct: 182 ASSKKSKKKQKAADEGDEMAVDKPT-VTNGDAVEDHKSEKKKKKKKKKR 325
>TC19835 similar to UP|Q9ZRW0 (Q9ZRW0) Nucleolar protein (Fragment), partial
(16%)
Length = 550
Score = 122 bits (306), Expect = 1e-28
Identities = 75/117 (64%), Positives = 87/117 (74%), Gaps = 8/117 (6%)
Frame = +1
Query: 344 GVAPRKNIDVMKSAIESVDNKDTEMETEEV----SAKKTKKKKQKAADGDDMAVDKAAEI 399
GVAPRKNIDVMKSAIES N D EMETEEV S+KK+KKK++ A +GD+MAVDK +
Sbjct: 1 GVAPRKNIDVMKSAIESAANNDAEMETEEVPVEASSKKSKKKQKAADEGDEMAVDKPT-V 177
Query: 400 TNGDA-EDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANADSSKK---KKKKSKKKD 452
TNGDA EDHKSE KKKKKEKRKLDQEV E+G N ++S+ KKKK +K D
Sbjct: 178 TNGDAVEDHKSE-KKKKKEKRKLDQEV-----AAENGTNGNASEPRSVKKKKDRKDD 330
>TC11310 similar to UP|BAD11331 (BAD11331) BRI1-KD interacting protein 102
(Fragment), partial (13%)
Length = 667
Score = 50.8 bits (120), Expect = 5e-07
Identities = 33/99 (33%), Positives = 48/99 (48%), Gaps = 16/99 (16%)
Frame = +1
Query: 372 EVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKK----------------K 415
EV +K +KKK+K D ++ E+ + D E + EKKKK K
Sbjct: 67 EVEGEKKEKKKKKKKDQEN------GEVASSDEEKAEKEKKKKHKVKVEDGSPDLDQSEK 228
Query: 416 KEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSKKKDAE 454
K+K+K DQ+ E + +G S+KK KK K +DAE
Sbjct: 229 KKKKKKDQDAEDNAAEISNGKEDRKSEKKHKKKKNQDAE 345
Score = 48.1 bits (113), Expect = 3e-06
Identities = 35/98 (35%), Positives = 54/98 (54%)
Frame = +1
Query: 327 GEKLREQVEERLDFYDKGVAPRKNIDVMKSAIESVDNKDTEMETEEVSAKKTKKKKQKAA 386
GEK ++ +++ D + VA + D K+ E ++E +++KKK+K
Sbjct: 76 GEKKEKKKKKKKDQENGEVA---SSDEEKAEKEKKKKHKVKVEDGSPDLDQSEKKKKKKK 246
Query: 387 DGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQE 424
D D A D AAEI+NG ED KSEKK KKK+ + ++E
Sbjct: 247 DQD--AEDNAAEISNG-KEDRKSEKKHKKKKNQDAEEE 351
Score = 32.0 bits (71), Expect = 0.22
Identities = 25/67 (37%), Positives = 35/67 (51%), Gaps = 20/67 (29%)
Frame = +1
Query: 408 KSEKKKKKKEKRKL--------------DQEVEVEDK-----VVEDGA-NADSSKKKKKK 447
K E + +KKEK+K +++ E E K VEDG+ + D S+KKKKK
Sbjct: 61 KVEVEGEKKEKKKKKKKDQENGEVASSDEEKAEKEKKKKHKVKVEDGSPDLDQSEKKKKK 240
Query: 448 SKKKDAE 454
K +DAE
Sbjct: 241 KKDQDAE 261
>TC12224 similar to PIR|D96602|D96602 nucleolar protein [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(16%)
Length = 315
Score = 44.3 bits (103), Expect = 4e-05
Identities = 20/22 (90%), Positives = 22/22 (99%)
Frame = +2
Query: 1 MTDELRTVLETNLPKVKEGKKA 22
+TDELRTVLETNLPKVKEGKK+
Sbjct: 248 LTDELRTVLETNLPKVKEGKKS 313
>BP031757
Length = 507
Score = 40.0 bits (92), Expect = 8e-04
Identities = 24/78 (30%), Positives = 39/78 (49%)
Frame = +2
Query: 365 DTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQE 424
D E ET K K+K+ K + D+ E +G+ + + EKKKKKK ++K ++E
Sbjct: 98 DEEEETPPQQLKPRKRKRMKLLEEDN-------EDKDGEDYEEEEEKKKKKKSRKKKEEE 256
Query: 425 VEVEDKVVEDGANADSSK 442
E E + E + +K
Sbjct: 257 EEEEQEEPESPQPPEDAK 310
Score = 37.0 bits (84), Expect = 0.007
Identities = 22/68 (32%), Positives = 35/68 (51%), Gaps = 2/68 (2%)
Frame = +2
Query: 389 DDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQ--EVEVEDKVVEDGANADSSKKKKK 446
+ M + ++ D E+ ++ ++ K KRK + E + EDK ED + KKKKK
Sbjct: 53 EKMVNRRFTQVATSDDEEEETPPQQLKPRKRKRMKLLEEDNEDKDGEDYEEEEEKKKKKK 232
Query: 447 KSKKKDAE 454
KKK+ E
Sbjct: 233 SRKKKEEE 256
>TC14320 similar to UP|Q41111 (Q41111) Dehydrin, partial (56%)
Length = 1231
Score = 38.5 bits (88), Expect = 0.002
Identities = 44/185 (23%), Positives = 80/185 (42%), Gaps = 19/185 (10%)
Frame = +3
Query: 286 HSSFIGRASARNKGRMARYLANKCSIASRIDCFS---EKGTSAFGEKLREQVEE---RLD 339
HSS ++ + + YL + + S I F K ++ E+ + + E ++
Sbjct: 228 HSSTSNSSNPSHLSQKLAYLLHTSVLISFISLFIFVISK*STIMAEENQNKYESGTTEVE 407
Query: 340 FYDKGV---------APRKNIDVMKSAIESVD-NKDTEMETEEVSAKKTKKKK---QKAA 386
D+GV + +V+ + E V +++ E + EEV + KK +K
Sbjct: 408 TQDRGVFDFLGKKKEEEKPQEEVIVTEFEKVKVSEEPEKKKEEVDHEGEKKHSTLLEKLH 587
Query: 387 DGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKK 446
D + + E + E+ K +KKK+KKE + D+ V +V D A+A+ K
Sbjct: 588 RSDSSSSSSSDEEEGKEGEEKKKKKKKEKKEVKSTDENTSVPVEV--DPAHAEEKKGFLD 761
Query: 447 KSKKK 451
K K+K
Sbjct: 762 KIKEK 776
Score = 32.0 bits (71), Expect = 0.22
Identities = 25/97 (25%), Positives = 42/97 (42%), Gaps = 8/97 (8%)
Frame = +3
Query: 366 TEMETEE------VSAKKTKKKKQKAADGDDMAVDKAAEITNGDAE--DHKSEKKKKKKE 417
TE+ET++ + KK ++K Q+ + K +E E DH+ EKK
Sbjct: 396 TEVETQDRGVFDFLGKKKEEEKPQEEVIVTEFEKVKVSEEPEKKKEEVDHEGEKKHSTLL 575
Query: 418 KRKLDQEVEVEDKVVEDGANADSSKKKKKKSKKKDAE 454
++ + E+ KKKKKK +KK+ +
Sbjct: 576 EKLHRSDSSSSSSSDEEEGKEGEEKKKKKKKEKKEVK 686
>AW720255
Length = 532
Score = 38.1 bits (87), Expect = 0.003
Identities = 20/61 (32%), Positives = 33/61 (53%)
Frame = +2
Query: 370 TEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVED 429
T + K TK++++ DG+ +D+ +NG+ KKK K K+KL++E E D
Sbjct: 119 TTQTPTKNTKRRRRTMQDGE---IDQTTTTSNGENNSESKSKKKLKM-KKKLEKETEKAD 286
Query: 430 K 430
K
Sbjct: 287 K 289
Score = 26.9 bits (58), Expect = 7.0
Identities = 14/41 (34%), Positives = 21/41 (51%), Gaps = 2/41 (4%)
Frame = +2
Query: 413 KKKKEKRKLDQEVEVEDKVVEDGA--NADSSKKKKKKSKKK 451
K K +R+ Q+ E++ N++S KKK K KKK
Sbjct: 137 KNTKRRRRTMQDGEIDQTTTTSNGENNSESKSKKKLKMKKK 259
>TC19735 similar to UP|Q8NJM5 (Q8NJM5) Probable ATP-dependent RNA helicase,
partial (5%)
Length = 565
Score = 38.1 bits (87), Expect = 0.003
Identities = 23/60 (38%), Positives = 35/60 (58%), Gaps = 8/60 (13%)
Frame = +3
Query: 403 DAEDHKSEKKKKKKEKRKLDQEVEVEDKVVED----GANAD----SSKKKKKKSKKKDAE 454
+ E++K +K+KK K +K ++V E+ E+ G N+D SSKKKKKK K + E
Sbjct: 159 ETENNKKKKEKKNKHDKKRPRDVANEEHAEEEEEQIGNNSDRDGESSKKKKKKKHKVEVE 338
Score = 37.0 bits (84), Expect = 0.007
Identities = 29/100 (29%), Positives = 43/100 (43%), Gaps = 17/100 (17%)
Frame = +3
Query: 344 GVAPRKNIDVMKSAIESVDN-----------KDTEMETEEVSAKKTKKKKQKAADGDDMA 392
GV P + ++ E+V N +ETE KK KK K D+A
Sbjct: 51 GVLPSPLNEAYETVAEAVQNLIMAEPEDNLQSSPMVETENNKKKKEKKNKHDKKRPRDVA 230
Query: 393 VDKAAEI------TNGDAEDHKSEKKKKKKEKRKLDQEVE 426
++ AE N D + S+KKKKKK K ++++ E
Sbjct: 231 NEEHAEEEEEQIGNNSDRDGESSKKKKKKKHKVEVEERKE 350
>BF177754
Length = 416
Score = 37.0 bits (84), Expect = 0.007
Identities = 24/90 (26%), Positives = 42/90 (46%)
Frame = +2
Query: 365 DTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQE 424
++ T+EV K +K+++K HK +KK K+K++ K
Sbjct: 2 NSRTRTDEVDLLKKEKRREKK---------------------HKKDKKDKEKKESK---- 106
Query: 425 VEVEDKVVEDGANADSSKKKKKKSKKKDAE 454
E +K +DG + + KK+K + KKKD +
Sbjct: 107 -EKREKDGKDGKHKEKEKKEKHRDKKKDKD 193
>TC13322 similar to UP|Q9ZS13 (Q9ZS13) RNA helicase (Fragment), partial (7%)
Length = 528
Score = 36.6 bits (83), Expect = 0.009
Identities = 24/76 (31%), Positives = 36/76 (46%), Gaps = 1/76 (1%)
Frame = +3
Query: 367 EMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVE 426
E+++E+ KK KK K K +G+ + + E + +KKKKKKEK E
Sbjct: 120 ELQSEK---KKKKKNKDKHQNGEKTSPKRKLEELDNQNGTESEKKKKKKKEKHDNKSEEN 290
Query: 427 VED-KVVEDGANADSS 441
E+ K NAD +
Sbjct: 291 AEETKEANGNGNADET 338
Score = 35.4 bits (80), Expect = 0.020
Identities = 25/79 (31%), Positives = 38/79 (47%), Gaps = 4/79 (5%)
Frame = +3
Query: 378 TKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDK----VVE 433
T +K A + D+A ++ + +SEKKKKKK K K + K ++
Sbjct: 51 TMGRKHDAVAASEPVADEAP-----NSPELQSEKKKKKKNKDKHQNGEKTSPKRKLEELD 215
Query: 434 DGANADSSKKKKKKSKKKD 452
+ +S KKKKKK +K D
Sbjct: 216 NQNGTESEKKKKKKKEKHD 272
>AW720001
Length = 542
Score = 33.9 bits (76), Expect = 0.057
Identities = 21/56 (37%), Positives = 28/56 (49%), Gaps = 2/56 (3%)
Frame = +3
Query: 399 ITNGDAEDHKSEKKK--KKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSKKKD 452
+ + D D EK+K KK+ K+ L + K D AD KKKK+ KKKD
Sbjct: 9 VGDSDGSDSGDEKEKPAKKEPKKDLPSKASTSKKKPRD---ADEDGKKKKQKKKKD 167
>AV415208
Length = 242
Score = 32.7 bits (73), Expect = 0.13
Identities = 15/45 (33%), Positives = 25/45 (55%)
Frame = +3
Query: 404 AEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKS 448
AE +EKK EK +++ + E K+ +D ++ KK+ KKS
Sbjct: 36 AEKKPAEKKPAAAEKAPAEKKPKAEKKISKDATGSEKKKKRTKKS 170
>TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;}, partial (44%)
Length = 700
Score = 32.7 bits (73), Expect = 0.13
Identities = 20/84 (23%), Positives = 36/84 (42%)
Frame = +2
Query: 362 DNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKRKL 421
D+ + E E E++ + ++ AA+ ++ + D E GD D+ +K KRK
Sbjct: 200 DDDEEEEEEEDLGTEYLVRRTVAAAEDEEASSDFEPEEDEGDDNDNDDGEKAGVPSKRKR 379
Query: 422 DQEVEVEDKVVEDGANADSSKKKK 445
+ D +DG D K+
Sbjct: 380 SDKDGSGDDDSDDGGEDDERPSKR 451
>AV411443
Length = 429
Score = 32.3 bits (72), Expect = 0.17
Identities = 21/64 (32%), Positives = 30/64 (46%), Gaps = 5/64 (7%)
Frame = +1
Query: 395 KAAEITNGDAEDHKSEKKKKK-----KEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSK 449
K+ + + D ED KS KK K +R DQ+ + + ED ++ K K K K
Sbjct: 1 KSLKSGSDDPEDSKSLKKVSKFGPESSNQRSGDQQKLASEFLKEDSGDSKKGKDKNSKKK 180
Query: 450 KKDA 453
KK A
Sbjct: 181 KKGA 192
>TC14310 homologue to UP|Q9M3H6 (Q9M3H6) Histone H2B, partial (92%)
Length = 502
Score = 32.3 bits (72), Expect = 0.17
Identities = 17/45 (37%), Positives = 27/45 (59%)
Frame = +3
Query: 406 DHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSKK 450
+ K +KK EK +++ + E K+ ++G S+KKKKKSKK
Sbjct: 75 EKKPAEKKPAAEKAPAEKKPKAEKKIAKEGG----SEKKKKKSKK 197
Score = 27.3 bits (59), Expect = 5.3
Identities = 15/44 (34%), Positives = 22/44 (49%)
Frame = +3
Query: 375 AKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEK 418
AK KK +K + +K + A++ SEKKKKK +K
Sbjct: 66 AKAEKKPAEKKPAAEKAPAEKKPKAEKKIAKEGGSEKKKKKSKK 197
>TC14318 similar to UP|O64739 (O64739) Ran-binding protein (AtRanBP1b),
partial (73%)
Length = 1099
Score = 32.0 bits (71), Expect = 0.22
Identities = 21/87 (24%), Positives = 43/87 (49%)
Frame = +1
Query: 360 SVDNKDTEMETEEVSAKKTKKKKQKAADGDDMAVDKAAEITNGDAEDHKSEKKKKKKEKR 419
S++N + MET + A+ K + + A+D ++K + DAE KK++
Sbjct: 487 SIENCKSFMETFQEVAESQKTEDKDASDAAAGLIEKLSVEEKADAE---------KKDEV 639
Query: 420 KLDQEVEVEDKVVEDGANADSSKKKKK 446
K + + E ++ + + AD+ KK ++
Sbjct: 640 KTESKAEEQEPASGEKSKADADKKDEE 720
>BP085186
Length = 388
Score = 31.6 bits (70), Expect = 0.28
Identities = 18/56 (32%), Positives = 31/56 (55%)
Frame = -2
Query: 393 VDKAAEITNGDAEDHKSEKKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKS 448
V+ +A IT E K +KKKKKK+K+K ++ + + + KK+K+K+
Sbjct: 300 VN*SARITRMGKEMKKKKKKKKKKKKKKKKKKKKQNQRKKRMKRKMSTPKKEKEKT 133
Score = 27.3 bits (59), Expect = 5.3
Identities = 18/44 (40%), Positives = 20/44 (44%)
Frame = -2
Query: 408 KSEKKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSKKK 451
K KKKKKK+K+K KKKKKK KKK
Sbjct: 267 KEMKKKKKKKKKK---------------------KKKKKKKKKK 199
Score = 26.9 bits (58), Expect = 7.0
Identities = 14/43 (32%), Positives = 24/43 (55%)
Frame = -2
Query: 408 KSEKKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSKK 450
K +KKKKKK+K+K ++ K ++ + +K+K S K
Sbjct: 249 KKKKKKKKKKKKKKKKKQNQRKKRMKRKMSTPKKEKEKTPSCK 121
>AW720070
Length = 484
Score = 31.6 bits (70), Expect = 0.28
Identities = 17/40 (42%), Positives = 25/40 (62%)
Frame = +2
Query: 410 EKKKKKKEKRKLDQEVEVEDKVVEDGANADSSKKKKKKSK 449
+K+KKKK+KRK ++V+ E ++ KKKKKK K
Sbjct: 110 KKRKKKKKKRKKKKQVKKE-------SSQQQEKKKKKKKK 208
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.313 0.130 0.353
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,479,572
Number of Sequences: 28460
Number of extensions: 60424
Number of successful extensions: 994
Number of sequences better than 10.0: 77
Number of HSP's better than 10.0 without gapping: 470
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 728
length of query: 454
length of database: 4,897,600
effective HSP length: 93
effective length of query: 361
effective length of database: 2,250,820
effective search space: 812546020
effective search space used: 812546020
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 57 (26.6 bits)
Medicago: description of AC135505.8


