

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
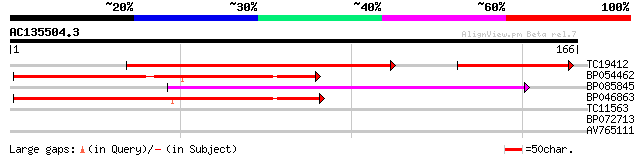
Query= AC135504.3 + phase: 0
(166 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC19412 similar to UP|Q84KB0 (Q84KB0) Pol protein, partial (7%) 96 8e-28
BP054462 73 2e-14
BP085845 63 2e-11
BP046863 58 7e-10
TC11563 27 1.8
BP072713 27 1.8
AV765111 25 6.9
>TC19412 similar to UP|Q84KB0 (Q84KB0) Pol protein, partial (7%)
Length = 519
Score = 95.5 bits (236), Expect(2) = 8e-28
Identities = 46/79 (58%), Positives = 61/79 (76%)
Frame = +3
Query: 35 RVTPMTGVRRALKSRKLTPKFIGPYQISERVGTVAYRVGLPTHLSNLHDVFHVSQLQKYV 94
RV+PM GV R K KL+P+FIGP+++ ERVG+V+YR+ LP LS +H VFHVS L+KY+
Sbjct: 3 RVSPMKGVLRFGKKGKLSPRFIGPFEVLERVGSVSYRLALPPDLSAVHPVFHVSMLRKYL 182
Query: 95 ADPSHVIPREDVQVRDNLT 113
DPSHVI EDVQ+ +L+
Sbjct: 183 YDPSHVIRHEDVQLDVHLS 239
Score = 43.1 bits (100), Expect(2) = 8e-28
Identities = 18/34 (52%), Positives = 22/34 (63%)
Frame = +2
Query: 132 KDIPLVKVVWGGVTGESLTWELESKMRDSYPESF 165
KD+ VKV+W G +GE TWE E MR+ YP F
Sbjct: 296 KDVGSVKVLWRGPSGEEATWEAEDIMREKYPHLF 397
>BP054462
Length = 422
Score = 73.2 bits (178), Expect = 2e-14
Identities = 38/93 (40%), Positives = 60/93 (63%), Gaps = 3/93 (3%)
Frame = +1
Query: 2 IREKMKASHSRQKSYHDKRRKALEFQEGDHVFLRVTPMTGVRRALKSR---KLTPKFIGP 58
+R + S ++Y +K+R+ +++Q GD VFL++ P RR+L + KL+P++ GP
Sbjct: 151 LRANLLKSQDMMRTYANKKRRDVDYQIGDEVFLKLQPYR--RRSLAKKMNEKLSPRYYGP 324
Query: 59 YQISERVGTVAYRVGLPTHLSNLHDVFHVSQLQ 91
Y I ++G VAYR+ LP H S +H VFHVS L+
Sbjct: 325 YPIVAKIGAVAYRLELPAH-SRVHPVFHVSLLK 420
>BP085845
Length = 464
Score = 63.2 bits (152), Expect = 2e-11
Identities = 37/106 (34%), Positives = 59/106 (54%)
Frame = -2
Query: 47 KSRKLTPKFIGPYQISERVGTVAYRVGLPTHLSNLHDVFHVSQLQKYVADPSHVIPREDV 106
K RK K IGP++I E V +AY + +L H V HV+ L + + D S I E V
Sbjct: 421 KKRKTESKIIGPFKILEMVCLIAY*LTHSLYLLAAHIVLHVTLLWRNLYDQSQNICHEGV 242
Query: 107 QVRDNLTVETSPLRIEDREVKKLRGKDIPLVKVVWGGVTGESLTWE 152
+ ++ + P+ + D +V+ +R K+I VKV+W G++G +WE
Sbjct: 241 XLGEHWSHMEHPIVMVDMKVRCMRPKNIDDVKVIWRGLSG*EKSWE 104
>BP046863
Length = 580
Score = 58.2 bits (139), Expect = 7e-10
Identities = 32/92 (34%), Positives = 58/92 (62%), Gaps = 1/92 (1%)
Frame = +1
Query: 2 IREKMKASHSRQKSYHDKRRKALEFQEGDHVFLRVTPMTGVRRAL-KSRKLTPKFIGPYQ 60
+R + + + ++ +K R+ +++Q G+ VFL++ P A K++KL+P+F GP++
Sbjct: 307 LRGNLLKAQDQMRAQANKHRRYVDYQVGNWVFLKLQPYKLQNLAQRKNQKLSPRFYGPFK 486
Query: 61 ISERVGTVAYRVGLPTHLSNLHDVFHVSQLQK 92
+ ERV VAY + L + S +H VFH+S L+K
Sbjct: 487 VLERVVQVAY*LDLXSE-SRVHPVFHLSLLEK 579
>TC11563
Length = 470
Score = 26.9 bits (58), Expect = 1.8
Identities = 11/30 (36%), Positives = 19/30 (62%)
Frame = +2
Query: 134 IPLVKVVWGGVTGESLTWELESKMRDSYPE 163
+P + + W G + TWEL S ++DS+P+
Sbjct: 8 VPQLLIQWEGAA--NCTWELLSYIQDSFPQ 91
>BP072713
Length = 403
Score = 26.9 bits (58), Expect = 1.8
Identities = 14/27 (51%), Positives = 16/27 (58%)
Frame = +3
Query: 42 VRRALKSRKLTPKFIGPYQISERVGTV 68
V + LKS K P+ G Y SERVG V
Sbjct: 162 VVQTLKSLKWPPRIPGYYDYSERVGVV 242
>AV765111
Length = 364
Score = 25.0 bits (53), Expect = 6.9
Identities = 18/57 (31%), Positives = 27/57 (46%)
Frame = +1
Query: 43 RRALKSRKLTPKFIGPYQISERVGTVAYRVGLPTHLSNLHDVFHVSQLQKYVADPSH 99
RR K T F PYQ+ + T+ + L +H L D ++ L KY+ P+H
Sbjct: 133 RRKTLVIKGTK*FYQPYQLFGCLKTITHCFYLDSHYGTLFDF*TLACLGKYI--PTH 297
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.135 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,461,374
Number of Sequences: 28460
Number of extensions: 27903
Number of successful extensions: 134
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 132
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 133
length of query: 166
length of database: 4,897,600
effective HSP length: 83
effective length of query: 83
effective length of database: 2,535,420
effective search space: 210439860
effective search space used: 210439860
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 52 (24.6 bits)
Medicago: description of AC135504.3


