

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135461.13 + phase: 0
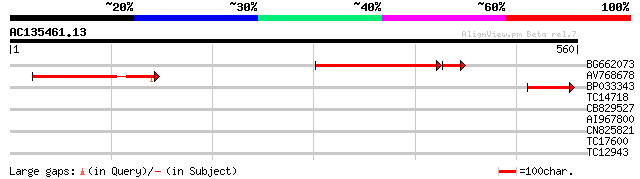
(560 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG662073 176 5e-51
AV768678 112 2e-25
BP033343 70 9e-13
TC14718 similar to UP|NCPR_CATRO (Q05001) NADPH--cytochrome P450... 30 1.0
CB829527 28 5.1
AI967800 27 8.7
CN825821 27 8.7
TC17600 similar to PIR|A47298|A47298 NADPH-ferrihemoprotein redu... 27 8.7
TC12943 similar to GB|AAO63388.1|28950929|BT005324 At5g19850 {Ar... 27 8.7
>BG662073
Length = 501
Score = 176 bits (445), Expect(2) = 5e-51
Identities = 74/124 (59%), Positives = 96/124 (76%)
Frame = +1
Query: 303 HGTNLPGGGYNDGAFWIDLPTDEDARSNLKKGNIESAELYVHVKPALGGAFTDIAMWVFC 362
+GTNLP + DGA+W+DLP D D + +KKG+++S E YVHVKP LGG FTD+AMWVF
Sbjct: 46 NGTNLPQDPHTDGAYWLDLPADADNKERVKKGDLQSFESYVHVKPMLGGTFTDLAMWVFY 225
Query: 363 PFNGPATLKVSLMNIEMNKIGEHVGDWEHFTLRVSNFTGELWSVFFSEHSGGKWVNAFDL 422
PFNGPA KV N+++ +IGEHVGDWEH TLR+SNF G+LW V+F++HS G WV++ +
Sbjct: 226 PFNGPARAKVEFFNVKLGRIGEHVGDWEHVTLRISNFDGQLWKVYFAQHSKGAWVDSSQI 405
Query: 423 EFIK 426
EF K
Sbjct: 406 EFQK 417
Score = 42.7 bits (99), Expect(2) = 5e-51
Identities = 17/23 (73%), Positives = 19/23 (81%)
Frame = +2
Query: 428 NKPIVYSSRHGHASYPHAGTYLQ 450
N+P+VYSS HGHASYPH G LQ
Sbjct: 431 NRPVVYSSLHGHASYPHPGLILQ 499
>AV768678
Length = 429
Score = 112 bits (280), Expect = 2e-25
Identities = 55/130 (42%), Positives = 80/130 (61%), Gaps = 4/130 (3%)
Frame = +3
Query: 23 FSLPSPLPQWPQGGGFAGGRISLGKIEVTKVNKFEKVWRCTNSNGKALGFTFYRPLEIPD 82
F LP+ +P WPQG GFA G I LG ++V++++ F+KVW G TF+ P E+P+
Sbjct: 63 FKLPANIPVWPQGDGFASGIIDLGDVKVSQISTFKKVWTTLEGGPDNAGATFFEPTELPE 242
Query: 83 GFSCLGYYCHSNDQPLRGHVLVARETTSKSQADCSESESPALKKPLNYSLIWCMDS---- 138
GF LG+Y N++P+ G VLVA+ D S S++ ALKKP++Y+L+W +S
Sbjct: 243 GFFTLGFYSQPNNKPVFGSVLVAK--------DDSPSDNEALKKPVDYTLVWSSNSKKIK 398
Query: 139 HDECVYFWLP 148
D+ Y WLP
Sbjct: 399 QDKDGYVWLP 428
>BP033343
Length = 481
Score = 70.1 bits (170), Expect = 9e-13
Identities = 30/47 (63%), Positives = 39/47 (82%)
Frame = -2
Query: 512 EIEKIIDMLPIFVRFSVENLFELFPTELSGEEGPTGPKEKDNWLGDE 558
E++KII+ LP ++ S++NLF FP EL GEEGPTGPKEK+NW+GDE
Sbjct: 480 ELDKIINALPKMLQHSMKNLFNQFPVELYGEEGPTGPKEKNNWIGDE 340
>TC14718 similar to UP|NCPR_CATRO (Q05001) NADPH--cytochrome P450 reductase
(CPR) (P450R) , partial (36%)
Length = 1163
Score = 30.0 bits (66), Expect = 1.0
Identities = 17/55 (30%), Positives = 25/55 (44%), Gaps = 1/55 (1%)
Frame = +1
Query: 252 LNQIHALIEHYGPTVYFHPDEKYMPSSVSW-FFKNGAILYTAGNAKGKAIDYHGT 305
L+++ GPT + + +S W GA +Y G+AKG A D H T
Sbjct: 511 LSELIVAFSREGPTKEYVQHKMMEKASDIWNMISQGAYIYVCGDAKGMARDVHRT 675
>CB829527
Length = 550
Score = 27.7 bits (60), Expect = 5.1
Identities = 17/39 (43%), Positives = 18/39 (45%)
Frame = -3
Query: 8 CWDSFPEFFDSDPLPFSLPSPLPQWPQGGGFAGGRISLG 46
CW P F LPF L S P+ GG GGR LG
Sbjct: 344 CW---PIFTLLHTLPFLLASKNTSHPKDGGGGGGRSILG 237
>AI967800
Length = 470
Score = 26.9 bits (58), Expect = 8.7
Identities = 14/42 (33%), Positives = 20/42 (47%), Gaps = 2/42 (4%)
Frame = +3
Query: 366 GPATLKVSLMNIEMNK--IGEHVGDWEHFTLRVSNFTGELWS 405
GP + ++NI K GE W+ F LRV +L+S
Sbjct: 249 GPVRIPTKVLNITTRKSPCGEGTNTWDRFELRVHKRVIDLYS 374
>CN825821
Length = 601
Score = 26.9 bits (58), Expect = 8.7
Identities = 14/42 (33%), Positives = 20/42 (47%), Gaps = 2/42 (4%)
Frame = +1
Query: 366 GPATLKVSLMNIEMNK--IGEHVGDWEHFTLRVSNFTGELWS 405
GP + ++NI K GE W+ F LRV +L+S
Sbjct: 214 GPVRMPTKVLNITTRKSPCGEGTNTWDRFELRVHKRVIDLYS 339
>TC17600 similar to PIR|A47298|A47298 NADPH-ferrihemoprotein reductase -
mung bean {Vigna radiata;} , partial (19%)
Length = 717
Score = 26.9 bits (58), Expect = 8.7
Identities = 12/20 (60%), Positives = 13/20 (65%)
Frame = +3
Query: 286 GAILYTAGNAKGKAIDYHGT 305
GA LY G+AKG A D H T
Sbjct: 237 GAYLYVCGDAKGMARDVHHT 296
>TC12943 similar to GB|AAO63388.1|28950929|BT005324 At5g19850 {Arabidopsis
thaliana;}, partial (28%)
Length = 578
Score = 26.9 bits (58), Expect = 8.7
Identities = 13/31 (41%), Positives = 17/31 (53%)
Frame = +1
Query: 117 SESESPALKKPLNYSLIWCMDSHDECVYFWL 147
S S + A+ L S+ WC +H CVYF L
Sbjct: 310 SSSGASAIS*FLCKSVYWCCSAH*NCVYFSL 402
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.139 0.449
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,793,149
Number of Sequences: 28460
Number of extensions: 179944
Number of successful extensions: 1004
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 997
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1003
length of query: 560
length of database: 4,897,600
effective HSP length: 95
effective length of query: 465
effective length of database: 2,193,900
effective search space: 1020163500
effective search space used: 1020163500
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Medicago: description of AC135461.13


