

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135415.3 + phase: 0 /pseudo
(436 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
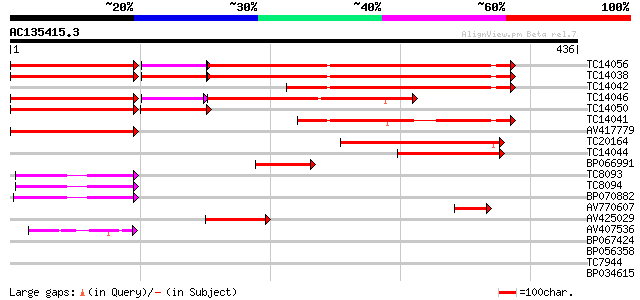
Score E
Sequences producing significant alignments: (bits) Value
TC14056 homologue to UP|P93769 (P93769) Elongation factor-1 alph... 308 9e-85
TC14038 homologue to UP|Q9LN13 (Q9LN13) T6D22.2, partial (47%) 303 3e-83
TC14042 homologue to UP|Q9XEW9 (Q9XEW9) Elongation factor 1-alph... 211 1e-55
TC14046 homologue to UP|P93769 (P93769) Elongation factor-1 alph... 201 2e-52
TC14050 homologue to UP|EF1A_TOBAC (P43643) Elongation factor 1-... 155 1e-50
TC14041 homologue to UP|Q9XEW9 (Q9XEW9) Elongation factor 1-alph... 162 1e-40
AV417779 145 1e-35
TC20164 UP|Q8LPC4 (Q8LPC4) Elongation factor 1-alpha (Elongation... 145 1e-35
TC14044 similar to UP|Q9LN13 (Q9LN13) T6D22.2, partial (10%) 107 3e-24
BP066991 55 2e-08
TC8093 homologue to UP|EFTU_TOBAC (P41342) Elongation factor Tu,... 51 4e-07
TC8094 homologue to UP|EFTU_PEA (O24310) Elongation factor Tu, c... 50 6e-07
BP070882 49 2e-06
AV770607 48 3e-06
AV425029 45 3e-05
AV407536 42 2e-04
BP067424 40 0.001
BP056358 39 0.002
TC7944 homologue to UP|Q9ASR1 (Q9ASR1) At1g56070/T6H22_13 (Elong... 36 0.011
BP034615 36 0.011
>TC14056 homologue to UP|P93769 (P93769) Elongation factor-1 alpha, complete
Length = 1897
Score = 308 bits (790), Expect = 9e-85
Identities = 157/237 (66%), Positives = 187/237 (78%)
Frame = +1
Query: 153 LQQAIDQISEPKRPEGKPLRLPLQHVYKIGGIGTVPVGRVETGVLKPGMVLTFAPTKLQS 212
L +A+DQI+EPKRP KPLRLPLQ VYKIGGIGTVPVGRVETGVLKPG V+TFAPT L +
Sbjct: 754 LLEALDQINEPKRPSDKPLRLPLQDVYKIGGIGTVPVGRVETGVLKPGFVVTFAPTGLTT 933
Query: 213 GVKSIQKFQENINEALPADIVGFRLTNKTLSDKNLMRGYVASNSEDEPAMEAAKFTSRVI 272
VKS++ E++ EALP D VGF + N + D L RG+VASNS+D+PA EAA FTS+VI
Sbjct: 934 EVKSVEMHHESLTEALPGDNVGFNVKNVAVKD--LKRGFVASNSKDDPAKEAANFTSQVI 1107
Query: 273 IINHPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPKSLKNGDAGIVKMI 332
I+NHP I GY P+LDCHTSH+AVKFA+L TK DR +G E+EK+PK LKNGDAG+VKM+
Sbjct: 1108IMNHPGQIGNGYAPVLDCHTSHIAVKFAELLTKIDRRSGKEIEKEPKFLKNGDAGMVKMV 1287
Query: 333 PMKPMVVEGINEYPSLGRFAVRDMRQTVAVGVIMSVKKNGYKNKAATKMTGSTTDQK 389
P KPMVVE +EYP LGRFAVRDMRQTVAVGVI SV+K K+ K+T + +K
Sbjct: 1288PTKPMVVETFSEYPPLGRFAVRDMRQTVAVGVIKSVEK---KDPTGAKVTKAAQKKK 1449
Score = 155 bits (393), Expect(2) = 9e-50
Identities = 77/99 (77%), Positives = 83/99 (83%)
Frame = +1
Query: 1 MVNSKPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVL 60
M K +INIV IGHV+SGKSTT GHL+YKLGGI K IE KEAAEM RSFKYAWVL
Sbjct: 109 MGKEKFHINIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVL 288
Query: 61 DKLKAERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
DKLKAERERGITIDI++ KFETTKY CT+IDAPGHRDFI
Sbjct: 289 DKLKAERERGITIDIALWKFETTKYYCTVIDAPGHRDFI 405
Score = 58.2 bits (139), Expect(2) = 9e-50
Identities = 27/54 (50%), Positives = 32/54 (59%)
Frame = +3
Query: 102 EHDHRDLRS*LCCSSHQFHYSRFASWLV*GWSDLGTCSTCVHSWCEANDLLLQQ 155
EHD+ + *LCC H FH F SW GW+D + C HSW +ANDLLL Q
Sbjct: 408 EHDYWNFSG*LCCPYH*FHNWWFRSWYFQGWTDS*ARTACFHSWSQANDLLL*Q 569
>TC14038 homologue to UP|Q9LN13 (Q9LN13) T6D22.2, partial (47%)
Length = 1844
Score = 303 bits (777), Expect = 3e-83
Identities = 153/237 (64%), Positives = 185/237 (77%)
Frame = +3
Query: 153 LQQAIDQISEPKRPEGKPLRLPLQHVYKIGGIGTVPVGRVETGVLKPGMVLTFAPTKLQS 212
L +A+DQI+EPKRP KPLRLPLQ VYKIGGIGTVPVGRVETG++KPGMV+TF PT L +
Sbjct: 735 LLEALDQINEPKRPSDKPLRLPLQDVYKIGGIGTVPVGRVETGLIKPGMVVTFGPTGLTT 914
Query: 213 GVKSIQKFQENINEALPADIVGFRLTNKTLSDKNLMRGYVASNSEDEPAMEAAKFTSRVI 272
VKS++ E + EALP D VGF + N + D L RG+VASNS+D+PA EAA FTS+VI
Sbjct: 915 EVKSVEMHHEALQEALPGDNVGFNVKNVAVKD--LKRGFVASNSKDDPAKEAANFTSQVI 1088
Query: 273 IINHPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPKSLKNGDAGIVKMI 332
I+NHP I GY P+LDCHTSH+AVKF ++ TK DR +G E+EK+PK LKNGDAG+VKM+
Sbjct: 1089IMNHPGQIGNGYAPVLDCHTSHIAVKFGEILTKIDRRSGKEIEKEPKFLKNGDAGMVKML 1268
Query: 333 PMKPMVVEGINEYPSLGRFAVRDMRQTVAVGVIMSVKKNGYKNKAATKMTGSTTDQK 389
P KPMVVE +EYP LGRFAVRDMRQTVAVGVI SV+K K+ K+T + +K
Sbjct: 1269PTKPMVVETFSEYPPLGRFAVRDMRQTVAVGVIKSVEK---KDPTGAKVTKAAAKKK 1430
Score = 156 bits (395), Expect(2) = 1e-51
Identities = 77/99 (77%), Positives = 83/99 (83%)
Frame = +3
Query: 1 MVNSKPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVL 60
M K +INIV IGHV+SGKSTT GHL+YKLGGI K IE KEAAEM RSFKYAWVL
Sbjct: 90 MGKEKVHINIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVL 269
Query: 61 DKLKAERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
DKLKAERERGITIDI++ KFETTKY CT+IDAPGHRDFI
Sbjct: 270 DKLKAERERGITIDIALWKFETTKYYCTVIDAPGHRDFI 386
Score = 63.5 bits (153), Expect(2) = 1e-51
Identities = 29/54 (53%), Positives = 36/54 (65%)
Frame = +2
Query: 102 EHDHRDLRS*LCCSSHQFHYSRFASWLV*GWSDLGTCSTCVHSWCEANDLLLQQ 155
EHD+ + *LCC+ H H+ F SW +*GW+D CS HSWCEA+DLLL Q
Sbjct: 389 EHDYWNFTG*LCCAHH*LHHWWF*SWYL*GWTDP*ACSPRFHSWCEADDLLL*Q 550
>TC14042 homologue to UP|Q9XEW9 (Q9XEW9) Elongation factor 1-alpha 1,
partial (38%)
Length = 800
Score = 211 bits (538), Expect = 1e-55
Identities = 108/176 (61%), Positives = 133/176 (75%)
Frame = +2
Query: 214 VKSIQKFQENINEALPADIVGFRLTNKTLSDKNLMRGYVASNSEDEPAMEAAKFTSRVII 273
VKS++ E + EALP D VGF + N + D L RG+VASNS+D+PA EAA FTS+VII
Sbjct: 5 VKSVEMHHEALQEALPGDNVGFNVKNVAVKD--LKRGFVASNSKDDPAKEAANFTSQVII 178
Query: 274 INHPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVELEKKPKSLKNGDAGIVKMIP 333
+NHP I GY P+LDCHTSH+AVKF+++ TK DR +G E+EK+PK LKNGDAG VKMIP
Sbjct: 179 MNHPGQIGNGYAPVLDCHTSHIAVKFSEILTKIDRRSGKEIEKEPKFLKNGDAGFVKMIP 358
Query: 334 MKPMVVEGINEYPSLGRFAVRDMRQTVAVGVIMSVKKNGYKNKAATKMTGSTTDQK 389
KPMVVE +EYP LGRFAVRDMRQTVAVGVI SV+K K+ + K+T + +K
Sbjct: 359 TKPMVVETFSEYPPLGRFAVRDMRQTVAVGVIKSVEK---KDPSGAKVTKAALKKK 517
>TC14046 homologue to UP|P93769 (P93769) Elongation factor-1 alpha, partial
(84%)
Length = 1506
Score = 201 bits (512), Expect = 2e-52
Identities = 101/163 (61%), Positives = 128/163 (77%), Gaps = 2/163 (1%)
Frame = +1
Query: 153 LQQAIDQISEPKRPEGKPLRLPLQHVYKIGGIGTVPVGRVETGVLKPGMVLTFAPTKLQS 212
L +A+DQI++PKRP KPLRLPLQ VYKIGGIGTVPVGRVETGVLKPG++++F P L +
Sbjct: 727 LLEALDQINKPKRPTDKPLRLPLQDVYKIGGIGTVPVGRVETGVLKPGVLVSFGPNGLTT 906
Query: 213 GVKSIQKFQENINEALPADIVGFRLTNKTLSDKNLMRGYVASNSEDEPAMEAAKFTSRVI 272
VKS++ E + EALP D VGF + K ++ K+L RG+VASNS+D+PA EAA FTS+VI
Sbjct: 907 EVKSMEMHHEALQEALPGDNVGFHV--KNVAVKDLKRGFVASNSKDDPAKEAANFTSQVI 1080
Query: 273 IINHPSVITKGYTPI--LDCHTSHVAVKFAKLATKFDRETGVE 313
I+NHP I GY P+ LDCHTSH+AVKF+++ TK DR +G E
Sbjct: 1081IMNHPGQIGNGYAPVLDLDCHTSHIAVKFSEILTKIDRRSGKE 1209
Score = 154 bits (388), Expect(2) = 2e-46
Identities = 76/99 (76%), Positives = 82/99 (82%)
Frame = +1
Query: 1 MVNSKPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVL 60
M K +INIV IGHV+SGKSTT GHL+YKLGGI K IE KEAAEM RSFKYAWVL
Sbjct: 82 MGKEKFHINIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVL 261
Query: 61 DKLKAERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
DKLKAERERGITIDI++ KFET KY CT+IDAPGHRDFI
Sbjct: 262 DKLKAERERGITIDIALWKFETNKYYCTVIDAPGHRDFI 378
Score = 48.9 bits (115), Expect(2) = 2e-46
Identities = 24/52 (46%), Positives = 31/52 (59%)
Frame = +3
Query: 102 EHDHRDLRS*LCCSSHQFHYSRFASWLV*GWSDLGTCSTCVHSWCEANDLLL 153
EHDHR++ LC S + FH F SW GW+D S C+HS + +DLLL
Sbjct: 381 EHDHRNIPGRLCRSYY*FHNWWF*SWYFQGWTDS*ARSPCIHSRSQTDDLLL 536
>TC14050 homologue to UP|EF1A_TOBAC (P43643) Elongation factor 1-alpha
(EF-1-alpha) (Vitronectin-like adhesion protein 1)
(PVN1), partial (38%)
Length = 583
Score = 155 bits (393), Expect(2) = 1e-50
Identities = 77/99 (77%), Positives = 83/99 (83%)
Frame = +2
Query: 1 MVNSKPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVL 60
M K +INIV IGHV+SGKSTT GHL+YKLGGI K IE KEAAEM RSFKYAWVL
Sbjct: 71 MGKEKFHINIVVIGHVDSGKSTTTGHLIYKLGGIDKRVIERFEKEAAEMNKRSFKYAWVL 250
Query: 61 DKLKAERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
DKLKAERERGITIDI++ KFETTKY CT+IDAPGHRDFI
Sbjct: 251 DKLKAERERGITIDIALWKFETTKYYCTVIDAPGHRDFI 367
Score = 60.8 bits (146), Expect(2) = 1e-50
Identities = 30/55 (54%), Positives = 39/55 (70%)
Frame = +1
Query: 101 *EHDHRDLRS*LCCSSHQFHYSRFASWLV*GWSDLGTCSTCVHSWCEANDLLLQQ 155
*E+D+ D+ S*LCC+ + FHY F S GW+D TCS+ +SWCEA+DLLL Q
Sbjct: 367 *EYDYWDIPS*LCCAHY*FHYWWF*SRYFQGWTDS*TCSSSFYSWCEADDLLL*Q 531
>TC14041 homologue to UP|Q9XEW9 (Q9XEW9) Elongation factor 1-alpha 1,
partial (34%)
Length = 777
Score = 162 bits (410), Expect = 1e-40
Identities = 91/170 (53%), Positives = 111/170 (64%), Gaps = 2/170 (1%)
Frame = +1
Query: 222 ENINEALPADIVGFRLTNKTLSDKNLMRGYVASNSEDEPAMEAAKFTSRVIIINHPSVIT 281
E + EALP D VGF + N + D L RG+VASNS+D+PA EAA FTS+VII+NHP I
Sbjct: 13 EALQEALPGDNVGFHVKNVAVKD--LKRGFVASNSKDDPAQEAANFTSQVIIMNHPGQIG 186
Query: 282 KGYTPILD--CHTSHVAVKFAKLATKFDRETGVELEKKPKSLKNGDAGIVKMIPMKPMVV 339
GY P+LD CHTSH+AVKF+++ TK DR +G VKMIP KPMVV
Sbjct: 187 NGYAPVLDLDCHTSHIAVKFSEILTKIDRRSG----------------FVKMIPTKPMVV 318
Query: 340 EGINEYPSLGRFAVRDMRQTVAVGVIMSVKKNGYKNKAATKMTGSTTDQK 389
E +EYP LGRFAVRDMRQTV GVI SV+K K + K+T + +K
Sbjct: 319 ETFSEYPLLGRFAVRDMRQTVVTGVIKSVEK---KEPSGAKVTKAALKKK 459
>AV417779
Length = 401
Score = 145 bits (367), Expect = 1e-35
Identities = 72/99 (72%), Positives = 81/99 (81%)
Frame = +1
Query: 1 MVNSKPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVL 60
M K +++IV IGHV+SGKSTT GHL+YK GGI+K AIE KEAAEM SFKYAWVL
Sbjct: 79 MGKEKQHVSIVVIGHVDSGKSTTTGHLIYKCGGIEKRAIEKFEKEAAEMGKGSFKYAWVL 258
Query: 61 DKLKAERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
DKLKAERERGITIDI++ KFET KY T+IDAPGHRDFI
Sbjct: 259 DKLKAERERGITIDIALWKFETEKYSFTIIDAPGHRDFI 375
>TC20164 UP|Q8LPC4 (Q8LPC4) Elongation factor 1-alpha (Elongation
factor-1a), partial (29%)
Length = 631
Score = 145 bits (366), Expect = 1e-35
Identities = 71/129 (55%), Positives = 92/129 (71%), Gaps = 3/129 (2%)
Frame = +3
Query: 255 NSEDEPAMEAAKFTSRVIIINHPSVITKGYTPILDCHTSHVAVKFAKLATKFDRETGVEL 314
+S+++P A F ++VII+NHP I GY P+LDCHT+H+A KF++L K DR +G +L
Sbjct: 9 DSKNDPPKGCASFNAQVIILNHPGEIHAGYAPVLDCHTAHIACKFSELILKMDRRSGKKL 188
Query: 315 EKKPKSLKNGDAGIVKMIPMKPMVVEGINEYPSLGRFAVRDMRQTVAVGVIMSVKK---N 371
E PK +K+GDA +VKM+ KPM VE +YP LGRFAVRDMRQTVAVGVI SV+K
Sbjct: 189 EDTPKMIKSGDAAMVKMVASKPMCVEAFTQYPPLGRFAVRDMRQTVAVGVIKSVEKKEVE 368
Query: 372 GYKNKAATK 380
G K+A K
Sbjct: 369 GKMTKSAAK 395
>TC14044 similar to UP|Q9LN13 (Q9LN13) T6D22.2, partial (10%)
Length = 550
Score = 107 bits (268), Expect = 3e-24
Identities = 55/82 (67%), Positives = 63/82 (76%)
Frame = +2
Query: 299 FAKLATKFDRETGVELEKKPKSLKNGDAGIVKMIPMKPMVVEGINEYPSLGRFAVRDMRQ 358
F +L TK DR +G ELEK+P+ LKNGDAG+VKMIP +PMVVE +EYP LGRFAVRDMRQ
Sbjct: 2 FGELLTKIDRRSG*ELEKEPQFLKNGDAGLVKMIPTQPMVVETFSEYPPLGRFAVRDMRQ 181
Query: 359 TVAVGVIMSVKKNGYKNKAATK 380
TVAVGVI SV+K TK
Sbjct: 182 TVAVGVIKSVEKKDPTGAKVTK 247
>BP066991
Length = 458
Score = 55.5 bits (132), Expect = 2e-08
Identities = 26/46 (56%), Positives = 34/46 (73%)
Frame = -2
Query: 190 GRVETGVLKPGMVLTFAPTKLQSGVKSIQKFQENINEALPADIVGF 235
GRVETGVLKPG++++F P L + VKS++ E + EALP D VGF
Sbjct: 457 GRVETGVLKPGVLVSFGPNGLTTEVKSMEMHHEALQEALPGDNVGF 320
>TC8093 homologue to UP|EFTU_TOBAC (P41342) Elongation factor Tu,
chloroplast precursor (EF-Tu), partial (58%)
Length = 1146
Score = 50.8 bits (120), Expect = 4e-07
Identities = 30/95 (31%), Positives = 45/95 (46%)
Frame = +1
Query: 5 KPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVLDKLK 64
KP++NI IGHV+ GK+T L L + SA + + +D
Sbjct: 346 KPHVNIGTIGHVDHGKTTLTAALTMALASLGNSAPKKYDE---------------IDAAP 480
Query: 65 AERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
ER RGITI+ + ++ET +D PGH D++
Sbjct: 481 EERARGITINTATVEYETENRHYAHVDCPGHADYV 585
Score = 35.8 bits (81), Expect = 0.014
Identities = 23/82 (28%), Positives = 36/82 (43%), Gaps = 5/82 (6%)
Frame = +1
Query: 143 HSWCEANDLLLQQAIDQISEPKRPEGKPLRLPLQHVYKIGGIGTVPVGRVETGVLKPGMV 202
+ W + L+ I P+R P ++ V+ I G GTV GR+E G++K G
Sbjct: 895 NQWVDKIYELMDNVDSYIPIPQRQTDLPFLCAIEDVFSITGRGTVATGRIERGLVKVGDT 1074
Query: 203 LTF-----APTKLQSGVKSIQK 219
+ T +GV+ QK
Sbjct: 1075VEIVGVRETRTTTVTGVEMFQK 1140
>TC8094 homologue to UP|EFTU_PEA (O24310) Elongation factor Tu, chloroplast
precursor (EF-Tu), partial (60%)
Length = 1092
Score = 50.4 bits (119), Expect = 6e-07
Identities = 30/95 (31%), Positives = 45/95 (46%)
Frame = +1
Query: 5 KPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVLDKLK 64
KP++NI IGHV+ GK+T L L + SA + + +D
Sbjct: 319 KPHVNIGTIGHVDHGKTTLTAALTMALASLGNSAPKKYDE---------------IDAAP 453
Query: 65 AERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
ER RGITI+ + ++ET +D PGH D++
Sbjct: 454 EERARGITINTATVEYETETRHYAHVDCPGHADYV 558
Score = 38.5 bits (88), Expect = 0.002
Identities = 20/61 (32%), Positives = 30/61 (48%)
Frame = +1
Query: 143 HSWCEANDLLLQQAIDQISEPKRPEGKPLRLPLQHVYKIGGIGTVPVGRVETGVLKPGMV 202
+ W + L+ I P+R P L ++ V+ I G GTV GR+E G +K G V
Sbjct: 868 NDWVDKIYQLMDNVDSYIPIPQRQTDLPFLLAIEDVFSITGRGTVATGRIERGTIKVGDV 1047
Query: 203 L 203
+
Sbjct: 1048V 1050
>BP070882
Length = 496
Score = 48.9 bits (115), Expect = 2e-06
Identities = 28/96 (29%), Positives = 47/96 (48%)
Frame = +2
Query: 4 SKPNINIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVLDKL 63
+KP++N+ +GHV+ GK+T + L K+ A + +DK
Sbjct: 194 TKPHVNVGTMGHVDHGKTTLTAAITKVLADEGKAKAIAFDE---------------IDKA 328
Query: 64 KAERERGITIDISMCKFETTKYCCTLIDAPGHRDFI 99
E++RGITI + ++ET K +D PGH D++
Sbjct: 329 PEEKKRGITIATAHVEYETVKRHYAHVDCPGHADYV 436
>AV770607
Length = 447
Score = 48.1 bits (113), Expect = 3e-06
Identities = 23/28 (82%), Positives = 25/28 (89%)
Frame = -1
Query: 343 NEYPSLGRFAVRDMRQTVAVGVIMSVKK 370
+EYP LGRFAVRDMRQTVAVGVI V+K
Sbjct: 441 SEYPPLGRFAVRDMRQTVAVGVIRRVEK 358
>AV425029
Length = 300
Score = 44.7 bits (104), Expect = 3e-05
Identities = 23/51 (45%), Positives = 32/51 (62%), Gaps = 1/51 (1%)
Frame = +1
Query: 151 LLLQQAIDQ-ISEPKRPEGKPLRLPLQHVYKIGGIGTVPVGRVETGVLKPG 200
L L A+D+ I +P R +P +P++ V+ I G GTV GRVE GV+K G
Sbjct: 139 LKLMDAVDEYIPDPVRQLDRPFLMPIEDVFSIQGRGTVATGRVEQGVIKVG 291
>AV407536
Length = 382
Score = 42.4 bits (98), Expect = 2e-04
Identities = 29/86 (33%), Positives = 43/86 (49%), Gaps = 2/86 (2%)
Frame = +2
Query: 15 HVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVLDKLKAERERGITID 74
H++SGK+T +L+ G I + E G++ KM D + ERE+GITI
Sbjct: 8 HIDSGKTTLTERVLFYAGRIHEIH-EVRGRDGVGAKM---------DSMDLEREKGITIQ 157
Query: 75 --ISMCKFETTKYCCTLIDAPGHRDF 98
+ C ++ K +ID PGH DF
Sbjct: 158SAATYCSWKDCK--INIIDTPGHVDF 229
>BP067424
Length = 473
Score = 39.7 bits (91), Expect = 0.001
Identities = 31/98 (31%), Positives = 43/98 (43%), Gaps = 1/98 (1%)
Frame = +1
Query: 2 VNSKPNI-NIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWVL 60
VNS+ I NI + HV+ GK+T A L+ G K A ++ +
Sbjct: 151 VNSRHKIRNICILAHVDHGKTTLADQLIAVASGGMVHP-----KVAGRVRF--------M 291
Query: 61 DKLKAERERGITIDISMCKFETTKYCCTLIDAPGHRDF 98
D L E+ R IT+ S + LID+PGH DF
Sbjct: 292 DYLDEEQRRAITMKSSSIALRYNHHTVNLIDSPGHIDF 405
>BP056358
Length = 598
Score = 38.5 bits (88), Expect = 0.002
Identities = 16/23 (69%), Positives = 20/23 (86%)
Frame = -1
Query: 324 GDAGIVKMIPMKPMVVEGINEYP 346
GDAG+VKM+P +PMVVE +EYP
Sbjct: 598 GDAGMVKMLPPQPMVVETFSEYP 530
>TC7944 homologue to UP|Q9ASR1 (Q9ASR1) At1g56070/T6H22_13 (Elongation
factor EF-2), complete
Length = 2971
Score = 36.2 bits (82), Expect = 0.011
Identities = 35/115 (30%), Positives = 51/115 (43%), Gaps = 17/115 (14%)
Frame = +1
Query: 1 MVNSKPNI-NIVFIGHVNSGKSTTAGHLLYKLGGIQKSAIEALGKEAAEMKMRSFKYAWV 59
+++ K NI N+ I HV+ GKST L+ G I + A +++M
Sbjct: 241 IMDLKHNIRNMSVIAHVDHGKSTLTDSLVAAAGIIAQEV-------AGDVRMT------- 378
Query: 60 LDKLKAERERGITI------------DISMCKFETTK----YCCTLIDAPGHRDF 98
D E ERGITI D+++ F+ + Y LID+PGH DF
Sbjct: 379 -DTRADEAERGITIKSTGISLYYEMTDVALKSFKGERMGNEYLINLIDSPGHVDF 540
>BP034615
Length = 287
Score = 36.2 bits (82), Expect = 0.011
Identities = 20/38 (52%), Positives = 26/38 (67%)
Frame = -2
Query: 352 AVRDMRQTVAVGVIMSVKKNGYKNKAATKMTGSTTDQK 389
AVRDMRQTVAVGVI SV+K K+ ++T + +K
Sbjct: 286 AVRDMRQTVAVGVIQSVEK---KDPTGAQVTQAAAQKK 182
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.321 0.136 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,765,710
Number of Sequences: 28460
Number of extensions: 93983
Number of successful extensions: 480
Number of sequences better than 10.0: 51
Number of HSP's better than 10.0 without gapping: 462
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 468
length of query: 436
length of database: 4,897,600
effective HSP length: 93
effective length of query: 343
effective length of database: 2,250,820
effective search space: 772031260
effective search space used: 772031260
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Medicago: description of AC135415.3


