

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135319.2 - phase: 0
(323 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
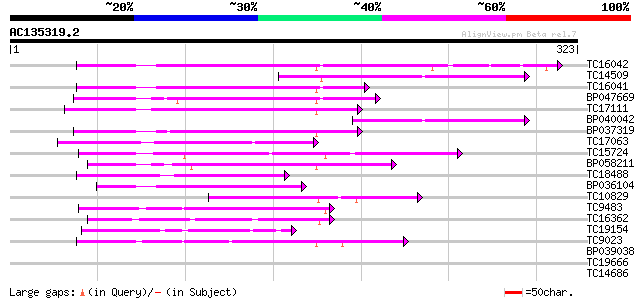
Score E
Sequences producing significant alignments: (bits) Value
TC16042 weakly similar to UP|Q8H0D9 (Q8H0D9) Alcohol dehydroge, ... 102 7e-23
TC14509 similar to UP|FABG_CUPLA (P28643) 3-oxoacyl-[acyl-carrie... 83 5e-17
TC16041 similar to UP|P93697 (P93697) CPRD12 protein, partial (60%) 79 8e-16
BP047669 79 8e-16
TC17111 weakly similar to GB|AAL99238.1|22651517|AY082345 short-... 73 6e-14
BP040042 71 3e-13
BP037319 70 6e-13
TC17063 weakly similar to UP|AAS50608 (AAS50608) ABL163Wp, parti... 62 2e-10
TC15724 similar to UP|Q9LHT0 (Q9LHT0) Short chain alcohol dehydr... 59 1e-09
BP058211 58 2e-09
TC18488 similar to UP|Q84JU0 (Q84JU0) Short chain alcohol dehydr... 58 2e-09
BP036104 44 3e-05
TC10829 similar to UP|Q93ZA0 (Q93ZA0) AT4g13250/F17N18_140, part... 43 8e-05
TC9483 similar to UP|Q9FK50 (Q9FK50) Brn1-like protein, partial ... 42 1e-04
TC16362 similar to UP|HCD2_HUMAN (Q99714) 3-hydroxyacyl-CoA dehy... 41 3e-04
TC19154 similar to UP|AAS38575 (AAS38575) Short-chain dehydrogen... 40 4e-04
TC9023 weakly similar to UP|Q9FXD7 (Q9FXD7) F12A21.14, partial (... 40 5e-04
BP039038 39 0.001
TC19666 38 0.002
TC14686 similar to UP|PORA_CUCSA (Q41249) Protochlorophyllide re... 38 0.003
>TC16042 weakly similar to UP|Q8H0D9 (Q8H0D9) Alcohol dehydroge, partial
(94%)
Length = 965
Score = 102 bits (255), Expect = 7e-23
Identities = 84/289 (29%), Positives = 145/289 (50%), Gaps = 12/289 (4%)
Frame = +2
Query: 39 KLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAK 98
+L+GK+A++TGG SGIG A LF+ GA V+ D D L + + +
Sbjct: 95 RLEGKVALITGGASGIGEATARLFSKHGAQVVIA-----------DIQDDLGHSVCKDLE 241
Query: 99 DPMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECG-SVEEIDEPRLERVFRT 157
+ ++ ++ + ++ ++ +G++DI+ NNA S+ + E+VF
Sbjct: 242 SASFVHCNVTKEDEVETAVNMAVSKHGKLDIMFNNAGISGNNNTSILNNTKSEFEQVFSV 421
Query: 158 NIFSYFFMTRHALKHM---KEGSNIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQ 214
N+ F T+HA + M + GS IINT S + G YTS+K A+V R +++
Sbjct: 422 NVSGAFLGTKHAARVMIPARRGS-IINTASTSGVIGGGAPHPYTSSKHAVVGLMRNTAVE 598
Query: 215 LVSKGIRVNGVAPGPIWTPLIPASF---NEEKTAQFGSDVPMKRAGQ-PVEVAPSFVFLA 270
L + G+RVN V+P I TP++ F E++ +F S+ +K A P +VA + ++L
Sbjct: 599 LEAYGVRVNCVSPYFIPTPMVKNFFKLGEEDEVPKFYSN--LKGADSVPEDVAEAVLYLG 772
Query: 271 SNQCSSYITGQVLHPNGDMLINASNVIYLFTHCM----VFLTHSFHFYF 315
S++ S Y++G L +G + +N +F + F+T FYF
Sbjct: 773 SDE-SKYVSGHNLVVDGGFTV-LNNGFCVFGQSV*NSYHFITCLLSFYF 913
>TC14509 similar to UP|FABG_CUPLA (P28643) 3-oxoacyl-[acyl-carrier protein]
reductase, chloroplast precursor (3-ketoacyl-acyl
carrier protein reductase) , partial (47%)
Length = 764
Score = 83.2 bits (204), Expect = 5e-17
Identities = 54/145 (37%), Positives = 77/145 (52%), Gaps = 2/145 (1%)
Frame = +3
Query: 154 VFRTNIFSYFFMTRHALKHMKEG--SNIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRAL 211
V N+ F T+ A K M + IIN SV GN +Y++ K ++ T+ +
Sbjct: 33 VIDINLTGVFLCTQAAAKIMMKNRKGRIINIASVVGLVGNVGQANYSAAKAGVIGLTKTV 212
Query: 212 SLQLVSKGIRVNGVAPGPIWTPLIPASFNEEKTAQFGSDVPMKRAGQPVEVAPSFVFLAS 271
+ + S+GI VN VAPG I + + A + + +P+ R GQP EVA FLA
Sbjct: 213 AKEYSSRGITVNAVAPGFIASDMT-AKLGNDIEKKILETIPLGRYGQPEEVAGLVEFLAL 389
Query: 272 NQCSSYITGQVLHPNGDMLINASNV 296
NQ +SYITGQVL +G M++ SNV
Sbjct: 390 NQAASYITGQVLTIDGGMVM*VSNV 464
>TC16041 similar to UP|P93697 (P93697) CPRD12 protein, partial (60%)
Length = 676
Score = 79.3 bits (194), Expect = 8e-16
Identities = 53/171 (30%), Positives = 89/171 (51%), Gaps = 4/171 (2%)
Frame = +2
Query: 39 KLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAK 98
+L+GK+A++TGG SGIG A LF+ GA V+ D D L + + +
Sbjct: 191 RLEGKVALITGGASGIGEATARLFSKHGAKVVIA-----------DIQDDLGHSVCRDLE 337
Query: 99 DPMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECG-SVEEIDEPRLERVFRT 157
+ ++ ++ + ++ ++ +G++DI+ NNA + S+ + E+VF
Sbjct: 338 SASFVHCNVTKEDEVETAVNTTVSKHGKLDIMFNNAGISGDNNISILNNTKSDFEQVFSV 517
Query: 158 NIFSYFFMTRHALKHM---KEGSNIINTTSVNAYKGNSTLIDYTSTKGAIV 205
N+F F T+HA + M + GS IINT SV+ G +T YTS+K A+V
Sbjct: 518 NVFGAFLGTKHAARVMVPARRGS-IINTASVSGLIGGATPHAYTSSKHAVV 667
>BP047669
Length = 544
Score = 79.3 bits (194), Expect = 8e-16
Identities = 56/181 (30%), Positives = 91/181 (49%), Gaps = 6/181 (3%)
Frame = +1
Query: 37 ANKLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKT-- 94
+ +L GK+A++TGG SGIG A LF GA VI D +D L M + KT
Sbjct: 28 SKRLDGKVAIITGGASGIGAATAKLFVQHGAKVIIA--------DIQDDLGM-SLCKTLE 180
Query: 95 ANAKDPMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECG-SVEEIDEPRLER 153
N + + D+ D + K +D ++ YG++DI+ NNA + S+ + +R
Sbjct: 181 PNFNNIIYAHCDVTNDSDVKNAVDMAVSKYGKLDIMYNNAGITGDLNLSILASSDECFKR 360
Query: 154 VFRTNIFSYFFMTRHALKHM---KEGSNIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRA 210
VF N++ F +HA + M K G I+ T+S+ + G YT++K A++ ++
Sbjct: 361 VFDVNVYGAFLGAKHAARVMIPAKRGV-ILFTSSIASILGGEAPHGYTASKHAVLGLMKS 537
Query: 211 L 211
L
Sbjct: 538 L 540
>TC17111 weakly similar to GB|AAL99238.1|22651517|AY082345 short-chain
dehydrogenase/reductase {Arabidopsis thaliana;}, partial
(53%)
Length = 561
Score = 73.2 bits (178), Expect = 6e-14
Identities = 49/173 (28%), Positives = 80/173 (45%), Gaps = 3/173 (1%)
Frame = +2
Query: 32 PDYKPANKLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKM 91
P PA +L GK+A+VTGG SGIG ++ LF GA + D +D L
Sbjct: 62 PAPAPAPRLLGKVALVTGGASGIGESIARLFHTNGAKICIA--------DMQDNLGNQVC 217
Query: 92 AKTANAKDPMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAA-EQYECGSVEEIDEPR 150
+ + + D+ + + + +D + +G +DI+VNNA C + +D
Sbjct: 218 ESLGDEANVCFLHCDVTEENDVRNAVDMTVAKFGTLDIIVNNAGISGAPCPDIRNVDLSD 397
Query: 151 LERVFRTNIFSYFFMTRHALKHM--KEGSNIINTTSVNAYKGNSTLIDYTSTK 201
++VF N+ F +HA + M K+ +II+ SV + G YT +K
Sbjct: 398 FDKVFNVNVKGVFHGMKHAARIMIPKKKGSIISLCSVASTMGGLGPHAYTGSK 556
>BP040042
Length = 512
Score = 70.9 bits (172), Expect = 3e-13
Identities = 40/101 (39%), Positives = 60/101 (58%)
Frame = -2
Query: 196 DYTSTKGAIVAFTRALSLQLVSKGIRVNGVAPGPIWTPLIPASFNEEKTAQFGSDVPMKR 255
+Y++ K ++ T+ ++ + S+GI VN VAPG I + + A + + +P+ R
Sbjct: 508 NYSAAKAGVIGLTKTVAKEYSSRGITVNAVAPGFIASDMT-AKLGNDIEKKILETIPLGR 332
Query: 256 AGQPVEVAPSFVFLASNQCSSYITGQVLHPNGDMLINASNV 296
GQP EVA FLA NQ +SYITGQVL +G M++ SNV
Sbjct: 331 YGQPEEVAGLVEFLALNQAASYITGQVLTIDGGMVM*VSNV 209
>BP037319
Length = 526
Score = 69.7 bits (169), Expect = 6e-13
Identities = 54/169 (31%), Positives = 80/169 (46%), Gaps = 4/169 (2%)
Frame = +2
Query: 37 ANKLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTAN 96
A +L+ K+AV+TGG GIG A LFA GA V+ D LD L A A
Sbjct: 56 AKRLEEKVAVITGGARGIGAATAKLFAENGAHVVIA-----------DVLDDLG-ASVAE 199
Query: 97 AKDPMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAA-EQYECGSVEEIDEPRLERVF 155
+ I D+ + + + I+ ++ G IDI+ NNA E S+ +D R+ RV
Sbjct: 200 SIGGRYIHCDVSKESDVESAINLAVSWKGHIDIMFNNAGIPDNEGRSISTLDMNRVNRVL 379
Query: 156 RTNIFSYFFMTRHALKHM---KEGSNIINTTSVNAYKGNSTLIDYTSTK 201
N++ +HA + M ++G +II T+S A G YT +K
Sbjct: 380 SINLYGTIHGIKHAARAMIKAQKGGSIICTSSAAATMGGFASHSYTMSK 526
>TC17063 weakly similar to UP|AAS50608 (AAS50608) ABL163Wp, partial (19%)
Length = 616
Score = 61.6 bits (148), Expect = 2e-10
Identities = 44/149 (29%), Positives = 68/149 (45%)
Frame = +3
Query: 28 QFTCPDYKPANKLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLD 87
QFT N L+GK+A++TGG SGIG + F GA+V + + LD
Sbjct: 189 QFTMESPFKGNILKGKVALITGGASGIGFEISTQFGKHGASVALMGRR-------KQVLD 347
Query: 88 MLKMAKTANAKDPMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECGSVEEID 147
+ + + D+ E+ RV++ +G+IDILVN AA + S E++
Sbjct: 348 SAVSVLQSLSIPAIGFVGDVRKQEDAARVVESTFKHFGKIDILVNAAAGNF-LVSAEDLS 524
Query: 148 EPRLERVFRTNIFSYFFMTRHALKHMKEG 176
V + F M ALK++K+G
Sbjct: 525 PNGFRTVLDIDSVGTFTMCSEALKYLKKG 611
>TC15724 similar to UP|Q9LHT0 (Q9LHT0) Short chain alcohol
dehydrogenase-like, partial (81%)
Length = 1014
Score = 58.9 bits (141), Expect = 1e-09
Identities = 59/227 (25%), Positives = 102/227 (43%), Gaps = 8/227 (3%)
Frame = +2
Query: 40 LQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAK- 98
L+G A+VTGG GIG AV A GATV +R+ +++ K K
Sbjct: 113 LKGTTALVTGGTRGIGHAVVEELAEFGATVYTC---------SRNEVELNACLKEWQEKG 265
Query: 99 -DPMAIPADLGFDENCKRVIDEIINAY-GRIDILVNNAAEQYECGSVEEIDEPRLERVFR 156
D +++ + + +A+ G+++ILVNN ++E E ++
Sbjct: 266 FSVSGSVCDASSPPQREKLFELVASAFNGKLNILVNNVGTNIRKPTIEYTAE-EYSKLMS 442
Query: 157 TNIFSYFFMTRHALKHMKEGSN----IINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALS 212
TN+ S +++ A +K N I++ + A+ G+ + Y ++K AI T+ L+
Sbjct: 443 TNLDSAHHLSQLAYPLLKASGNGSIVFISSVAALAHVGSGAV--YAASKAAINQLTKYLA 616
Query: 213 LQLVSKGIRVNGVAPGPIWTPLI-PASFNEEKTAQFGSDVPMKRAGQ 258
+ IR N VAP T L+ P ++E + S P+KR +
Sbjct: 617 CEWAKDNIRSNSVAPWYTKTSLVEPVLSHQELVNEILSRTPIKRMAE 757
>BP058211
Length = 562
Score = 58.2 bits (139), Expect = 2e-09
Identities = 48/181 (26%), Positives = 80/181 (43%), Gaps = 5/181 (2%)
Frame = +3
Query: 45 AVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAKDPMA-- 102
+++TG SGIG+A F GA VI D + L +TA P A
Sbjct: 54 SMITGAASGIGKAAATKFINNGAKVIIA--------DIQQKLGQ----ETAKELGPNATF 197
Query: 103 IPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYEC-GSVEEIDEPRLERVFRTNIFS 161
I D+ + + ++ I+ Y ++DI+ NNA GS+ ++D +RV N+
Sbjct: 198 ITCDVTKESDISNAVEFAISEYKQLDIMYNNAGVPCRTPGSIVDLDLASFDRVMDINVRG 377
Query: 162 YFFMTRHALKHM--KEGSNIINTTSVNAYKGNSTLIDYTSTKGAIVAFTRALSLQLVSKG 219
+HA + M + +I+ T SV G Y+ +K ++ ++LS +L G
Sbjct: 378 VMAGIKHAARVMIPRGAGSILCTASVTGVMGGLAQHTYSISKFTVIGIVKSLSSELCRHG 557
Query: 220 I 220
I
Sbjct: 558 I 560
>TC18488 similar to UP|Q84JU0 (Q84JU0) Short chain alcohol dehydrogenase
(Fragment), partial (47%)
Length = 440
Score = 58.2 bits (139), Expect = 2e-09
Identities = 32/121 (26%), Positives = 62/121 (50%)
Frame = +1
Query: 39 KLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAK 98
+ +GK+A+VT GIG ++ LEGA+V+ + K A + L A
Sbjct: 79 RFRGKVAIVTASTQGIGFSIAERLGLEGASVVISSRKQQNVDAAAEKL-------RAKGI 237
Query: 99 DPMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECGSVEEIDEPRLERVFRTN 158
D +A+ + + K +ID+ + YG+ID++V+NAA ++ + + L++++ N
Sbjct: 238 DVLAVVCHVSNAQQRKNLIDKTVQKYGKIDVVVSNAAANPSVDAILQTKDTVLDKLWEIN 417
Query: 159 I 159
+
Sbjct: 418 V 420
>BP036104
Length = 439
Score = 44.3 bits (103), Expect = 3e-05
Identities = 29/121 (23%), Positives = 54/121 (43%), Gaps = 1/121 (0%)
Frame = +2
Query: 50 GDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAKDPMAIPADLGF 109
G SGIG A LF+ GA V+ D D L + + K + ++
Sbjct: 107 GASGIGEATARLFSKHGAQVVIA-----------DVQDDLGQSLCKDLKSASFVHCNVTK 253
Query: 110 DENCKRVIDEIINAYGRIDILVNNAA-EQYECGSVEEIDEPRLERVFRTNIFSYFFMTRH 168
++ + ++ ++ +G++DI+ NNA ++ + + E+V TN+ F T+H
Sbjct: 254 EDEVETAVNTTVSKHGKLDIMFNNAGITGANKTNILDNTKSEFEQVINTNLVGVFLGTKH 433
Query: 169 A 169
A
Sbjct: 434 A 436
>TC10829 similar to UP|Q93ZA0 (Q93ZA0) AT4g13250/F17N18_140, partial (45%)
Length = 732
Score = 42.7 bits (99), Expect = 8e-05
Identities = 32/127 (25%), Positives = 60/127 (47%), Gaps = 5/127 (3%)
Frame = +1
Query: 114 KRVIDEIINAYGRIDILVNNAAEQYECGSVEEIDEPRLERVFRTNIFSYFFMTRHALKHM 173
+R+ + +N G IDI +NNA + E + ++++ TN+ TR A++ M
Sbjct: 298 QRLANFAVNELGYIDIWINNAGANKGFRPLLEFSDEDIQQIVSTNLVGSILCTREAMRIM 477
Query: 174 K---EGSNIINTTSVNAYKGNSTLID--YTSTKGAIVAFTRALSLQLVSKGIRVNGVAPG 228
+ + +I N + G+ST + Y STK + F +L + + V+ +PG
Sbjct: 478 RNQVKAGHIFNMDGAGS-GGSSTPLTAVYGSTKCGLRQFQGSLLKECKRSKVGVHTASPG 654
Query: 229 PIWTPLI 235
+ T L+
Sbjct: 655 MVLTDLL 675
Score = 26.9 bits (58), Expect = 4.7
Identities = 12/27 (44%), Positives = 16/27 (58%)
Frame = +2
Query: 46 VVTGGDSGIGRAVCNLFALEGATVIFT 72
V+TG G+G+A+ F L G VI T
Sbjct: 65 VITGSTRGLGKALAREFLLSGDRVIVT 145
>TC9483 similar to UP|Q9FK50 (Q9FK50) Brn1-like protein, partial (51%)
Length = 549
Score = 42.4 bits (98), Expect = 1e-04
Identities = 36/151 (23%), Positives = 66/151 (42%), Gaps = 5/151 (3%)
Frame = +2
Query: 40 LQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAKD 99
LQ ++A+VTG GIGR + A GA ++ Y + D +++ A +A ++
Sbjct: 107 LQDRVAIVTGSSRGIGREIAIHLASLGARLVINYT---SNSDQANSVAAQINAGSATSR- 274
Query: 100 PMAIPADLGFDENCKRVIDEIINAY-GRIDILVNNAAE-QYECGSVEEIDEPRLERVFRT 157
+ + AD+ + K + D A+ + ILVN+A ++ + +R+
Sbjct: 275 AITVKADVSDPDQVKSLFDSAEQAFDSPVHILVNSAGVIDSTYSTIADTTVETFDRIMSV 454
Query: 158 NIFSYFFMTRHALKHMKEGSN---IINTTSV 185
N F + +K G I+ TTS+
Sbjct: 455 NARGAFLCAKEGANRVKRGGGGRIILLTTSL 547
>TC16362 similar to UP|HCD2_HUMAN (Q99714) 3-hydroxyacyl-CoA dehydrogenase
type II (Type II HADH) (Endoplasmic
reticulum-associated amyloid beta-peptide binding
protein) (Short-chain type dehydrogenase/reductase
XH98G2) , partial (7%)
Length = 613
Score = 40.8 bits (94), Expect = 3e-04
Identities = 36/148 (24%), Positives = 64/148 (42%), Gaps = 7/148 (4%)
Frame = +2
Query: 45 AVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAKDPMAIP 104
A++TGG SGIG + +L VI + A++ ++ + D M +
Sbjct: 191 AIITGGASGIGLETARVLSLRKVHVI---IAARNMNSAKEAKQLILQDNESARVDIMKL- 358
Query: 105 ADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECGSVEEIDEPRLERVFRTNIFSYFF 164
DL ++ + ++ I ++IL+NNA + ++ E +E F TN +F
Sbjct: 359 -DLCSLKSVRSFVENFIALGLPLNILINNAGVMF---CPFQLSEDGIEMQFATNHLGHFL 526
Query: 165 MTRHALKHMKE-------GSNIINTTSV 185
+T L MK+ IIN +S+
Sbjct: 527 LTNLLLDKMKQTAKATGIXGRIINLSSI 610
>TC19154 similar to UP|AAS38575 (AAS38575) Short-chain dehydrogenase Tic32,
partial (44%)
Length = 508
Score = 40.4 bits (93), Expect = 4e-04
Identities = 38/122 (31%), Positives = 54/122 (44%)
Frame = +2
Query: 42 GKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAKDPM 101
G AVVTG SGIG + A G VI KD ++T+ LK +A
Sbjct: 164 GLTAVVTGASSGIGTETTRVLAKRGVHVIMGVRNTAAGKDVKETI--LKENPSAKVD--- 328
Query: 102 AIPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECGSVEEIDEPRLERVFRTNIFS 161
A+ DL E+ K+ E ++ ++IL+NNA C + D L+ F TN
Sbjct: 329 AMELDLSSMESVKKFASEYKSSGLPLNILINNAGIM-ACPFMLSKDNHELQ--FATNHLG 499
Query: 162 YF 163
+F
Sbjct: 500 HF 505
>TC9023 weakly similar to UP|Q9FXD7 (Q9FXD7) F12A21.14, partial (13%)
Length = 1282
Score = 40.0 bits (92), Expect = 5e-04
Identities = 44/194 (22%), Positives = 81/194 (41%), Gaps = 5/194 (2%)
Frame = +2
Query: 39 KLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKTANAK 98
K G AV+TG GIG+A+ A +G ++ G T ++ K
Sbjct: 155 KQYGSWAVITGSTDGIGKAMSFELASQGLNLLLI---GRNPTKLEATSKEIRDKYHVEVK 325
Query: 99 DPMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECGS-VEEIDEPRLERVFRT 157
+ + E ++I E I+ + ++VN+A Y + E+D ++ V +
Sbjct: 326 -ILVLDMQYVKGEEMGKLIVEAIDGLD-VGLVVNSAGLAYPYARFLHEVDAELMDAVVKV 499
Query: 158 NIFSYFFMTRHALKHM--KEGSNIINTTSVNAY--KGNSTLIDYTSTKGAIVAFTRALSL 213
N+ ++T+ L M K+ I+N S + + + Y +TK + F+R +SL
Sbjct: 500 NVEGLTWVTKAVLPGMIKKKKGAIVNIGSGSTFVLPSYPLVTLYAATKAYLAMFSRCISL 679
Query: 214 QLVSKGIRVNGVAP 227
+ GI + P
Sbjct: 680 EYKHHGIDIQCQIP 721
>BP039038
Length = 538
Score = 39.3 bits (90), Expect = 0.001
Identities = 40/150 (26%), Positives = 62/150 (40%), Gaps = 9/150 (6%)
Frame = +3
Query: 44 IAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDARDTLDMLKMAKT-----ANAK 98
+AVVTGG+ GIG A+ A G V+ T + A + L +A+ +
Sbjct: 84 VAVVTGGNRGIGFALVKRLAELGLNVVLTARNNQNGEAAMENLRAQGLAQNIHFLQLDVS 263
Query: 99 DPMAIPADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECGSVEEIDEPRLERVFRTN 158
DP +I A F + K +DILVNNA + + +D E V +TN
Sbjct: 264 DPASITA---FASSFKAQFGP------TLDILVNNAGVSFNDLNENSVD--HAESVIKTN 410
Query: 159 IFSYFFMTRHALKHMKE----GSNIINTTS 184
+ + L + G+ I+N +S
Sbjct: 411 FYGPKLLIEALLPLFRSSSSFGTRILNVSS 500
>TC19666
Length = 521
Score = 38.1 bits (87), Expect = 0.002
Identities = 23/63 (36%), Positives = 35/63 (55%), Gaps = 4/63 (6%)
Frame = +3
Query: 234 LIPASF----NEEKTAQFGSDVPMKRAGQPVEVAPSFVFLASNQCSSYITGQVLHPNGDM 289
L P+SF +++ + S P++R G P EV+ FL SSYITGQ++ +G M
Sbjct: 90 LSPSSFRFSAHKDYLEEVYSRTPLRRLGDPAEVSSLVAFLCL-PASSYITGQIICVDGGM 266
Query: 290 LIN 292
+N
Sbjct: 267 SVN 275
>TC14686 similar to UP|PORA_CUCSA (Q41249) Protochlorophyllide reductase,
chloroplast precursor (PCR) (NADPH-protochlorophyllide
oxidoreductase) (POR) , partial (51%)
Length = 706
Score = 37.7 bits (86), Expect = 0.003
Identities = 38/146 (26%), Positives = 59/146 (40%), Gaps = 2/146 (1%)
Frame = +2
Query: 24 NPTPQFTCPDYKPANKLQGKIAVVTGGDSGIGRAVCNLFALEGATVIFTYVKGHEDKDAR 83
+P + P+ K L+ V+TG SG+G A A G K H R
Sbjct: 296 SPAVNSSAPEAKKT--LRKGCVVITGASSGLGLATAKALAETG--------KWHVIMACR 445
Query: 84 DTLDMLKMAKTAN-AKDPMAI-PADLGFDENCKRVIDEIINAYGRIDILVNNAAEQYECG 141
D L + AK+A AK+ +I DL ++ ++ +D + +D+LV NAA
Sbjct: 446 DFLKAARAAKSAGIAKENYSIMHLDLSSLDSVRQFVDNFRRSEMPLDVLVCNAAVYLPTA 625
Query: 142 SVEEIDEPRLERVFRTNIFSYFFMTR 167
E TN +F ++R
Sbjct: 626 KEPTFTADGFELSVGTNHLGHFLLSR 703
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.135 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,127,773
Number of Sequences: 28460
Number of extensions: 66082
Number of successful extensions: 418
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 408
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 408
length of query: 323
length of database: 4,897,600
effective HSP length: 90
effective length of query: 233
effective length of database: 2,336,200
effective search space: 544334600
effective search space used: 544334600
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 55 (25.8 bits)
Medicago: description of AC135319.2


