

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
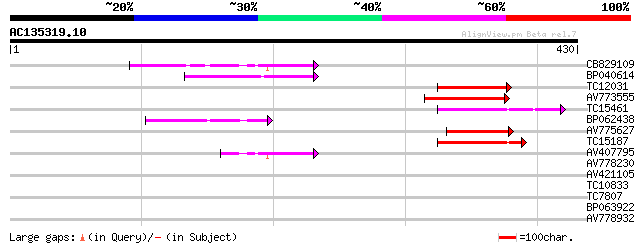
Query= AC135319.10 + phase: 1 /pseudo/partial
(430 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CB829109 83 8e-17
BP040614 73 1e-13
TC12031 homologue to UP|ATK1_ARATH (Q07970) Kinesin 1 (Kinesin-l... 68 3e-12
AV773555 64 5e-11
TC15461 similar to UP|O24147 (O24147) Kinesin-like protein, part... 63 1e-10
BP062438 60 9e-10
AV775627 58 3e-09
TC15187 similar to UP|O24147 (O24147) Kinesin-like protein, part... 54 4e-08
AV407795 48 3e-06
AV778230 37 0.006
AV421105 35 0.024
TC10833 similar to UP|Q84V30 (Q84V30) Phosphatidylserine decarbo... 27 5.0
TC7807 homologue to UP|Q946J9 (Q946J9) Aquaporin protein PIP1,1,... 27 6.5
BP063922 27 8.5
AV778932 27 8.5
>CB829109
Length = 514
Score = 83.2 bits (204), Expect = 8e-17
Identities = 56/145 (38%), Positives = 81/145 (55%), Gaps = 2/145 (1%)
Frame = +2
Query: 92 AQTKPIVASVLDGHNVCIFAYGQTGTGKTFTMEGTPEHRGVNYRTLEELFRVSEERQGTI 151
A +PI+ ++ + FAYGQTG+GKT+TM+ P + + +L R + Q
Sbjct: 5 ATVEPIIPTIFERTKATCFAYGQTGSGKTYTMQPLPLRAAEDL--VRQLHRPVYQNQ--- 169
Query: 152 KYELLVSMLEVYNEKIKDLLAGNSSEATKKLEVKQAADGTQEV--PGLVETHVYGADGVW 209
K++L +S E+Y K+ DLL+ KKL +++ DG Q+V GL E V+ V
Sbjct: 170 KFKLWLSYFEIYGGKLFDLLSDR-----KKLCMRE--DGRQQVCIVGLQEFEVFDVQIVK 328
Query: 210 EILKSGNRVRSVGSTSANELSSRSH 234
E ++ GN RS GST ANE SSRSH
Sbjct: 329 EFIEKGNAARSTGSTGANEESSRSH 403
>BP040614
Length = 504
Score = 72.8 bits (177), Expect = 1e-13
Identities = 39/102 (38%), Positives = 59/102 (57%)
Frame = +1
Query: 133 NYRTLEELFRVSEERQGTIKYELLVSMLEVYNEKIKDLLAGNSSEATKKLEVKQAADGTQ 192
NY L +LF +S++R +IKYE+ V M+E+YNE+++DLL N S + +G
Sbjct: 10 NY*GLRDLFHISKDRADSIKYEVFVQMIEIYNEQVRDLLVSNGSNRRLDIRNNSQLNGL- 186
Query: 193 EVPGLVETHVYGADGVWEILKSGNRVRSVGSTSANELSSRSH 234
VP V V +++ G + R+VG+T+ NE SSRSH
Sbjct: 187 NVPDAYLVPVTCTQDVLYLMEVGQKNRAVGATALNERSSRSH 312
>TC12031 homologue to UP|ATK1_ARATH (Q07970) Kinesin 1 (Kinesin-like protein
A), partial (7%)
Length = 740
Score = 68.2 bits (165), Expect = 3e-12
Identities = 34/56 (60%), Positives = 40/56 (70%)
Frame = +2
Query: 325 RNSKLTHILQSSLGGDCKTLMFVQISPSSVDLTETLCSLNFATRVRGIESGPARKQ 380
RNSKLT++LQ LGGD KTLMFV ISP + E+LCSL FA+RV E G R+Q
Sbjct: 2 RNSKLTYLLQPCLGGDSKTLMFVNISPDQSSVGESLCSLRFASRVNACEIGIPRRQ 169
>AV773555
Length = 460
Score = 63.9 bits (154), Expect = 5e-11
Identities = 32/65 (49%), Positives = 42/65 (64%)
Frame = -1
Query: 315 LPNQPTYLTDRNSKLTHILQSSLGGDCKTLMFVQISPSSVDLTETLCSLNFATRVRGIES 374
L Q ++ R+SKLT++LQ +LGGD KT+MFV ISP E+LCSL FA+RV
Sbjct: 427 LAQQDDHIPFRHSKLTYLLQPALGGDSKTVMFVNISPDPASAGESLCSLRFASRVNACVI 248
Query: 375 GPARK 379
G R+
Sbjct: 247 GTPRR 233
>TC15461 similar to UP|O24147 (O24147) Kinesin-like protein, partial (9%)
Length = 527
Score = 62.8 bits (151), Expect = 1e-10
Identities = 38/97 (39%), Positives = 58/97 (59%)
Frame = +3
Query: 325 RNSKLTHILQSSLGGDCKTLMFVQISPSSVDLTETLCSLNFATRVRGIESGPARKQVDLT 384
RN KLT ++ SLGG+ KTLMFV +SP L ET SL +A+RVR I + P+ K V
Sbjct: 51 RNHKLTMLMSDSLGGNAKTLMFVNVSPIESSLDETHNSLMYASRVRSIVNDPS-KNVSSK 227
Query: 385 ELLKYKQMAEKSKHDEKEARKLQDNLQSVQMRLATRE 421
E+++ K+ K ++ R +++L+ +Q T+E
Sbjct: 228 EIMRLKKQVAYWK-EQAGRRGEEEDLEEIQEVRPTKE 335
>BP062438
Length = 498
Score = 59.7 bits (143), Expect = 9e-10
Identities = 36/96 (37%), Positives = 56/96 (57%)
Frame = +3
Query: 104 GHNVCIFAYGQTGTGKTFTMEGTPEHRGVNYRTLEELFRVSEERQGTIKYELLVSMLEVY 163
G N +FAYGQT +GKT TM G+ GV R + +LF++ ++ ++ L +S +E+Y
Sbjct: 222 GFNGTVFAYGQTNSGKTHTMRGSKADPGVIPRAVRDLFQIIQQDVDR-EFLLRMSYMEIY 398
Query: 164 NEKIKDLLAGNSSEATKKLEVKQAADGTQEVPGLVE 199
NE+I DLLA +KL++ + + V GL E
Sbjct: 399 NEEINDLLAPEH----RKLQIHENLERGIYVAGLRE 494
>AV775627
Length = 448
Score = 58.2 bits (139), Expect = 3e-09
Identities = 29/51 (56%), Positives = 35/51 (67%)
Frame = -3
Query: 332 ILQSSLGGDCKTLMFVQISPSSVDLTETLCSLNFATRVRGIESGPARKQVD 382
I Q LGGD KTLMFV ISP ++E+LCSL FA+RV E G AR+Q +
Sbjct: 380 IEQPCLGGDSKTLMFVNISPDPSSVSESLCSLRFASRVNACEIGSARRQTN 228
>TC15187 similar to UP|O24147 (O24147) Kinesin-like protein, partial (8%)
Length = 611
Score = 54.3 bits (129), Expect = 4e-08
Identities = 30/68 (44%), Positives = 44/68 (64%)
Frame = +2
Query: 325 RNSKLTHILQSSLGGDCKTLMFVQISPSSVDLTETLCSLNFATRVRGIESGPARKQVDLT 384
RN KLT ++ SLGG+ +TLMFV +SP L ET SL +A+RVR I + P+ + V
Sbjct: 11 RNHKLTMLMSDSLGGNAQTLMFVNVSPVESSLDETHNSLMYASRVRSIINDPS-QNVASK 187
Query: 385 ELLKYKQM 392
E+ + K++
Sbjct: 188 EIARLKKL 211
>AV407795
Length = 424
Score = 48.1 bits (113), Expect = 3e-06
Identities = 32/76 (42%), Positives = 44/76 (57%), Gaps = 2/76 (2%)
Frame = +2
Query: 161 EVYNEKIKDLLAGNSSEATKKLEVKQAADGTQEV--PGLVETHVYGADGVWEILKSGNRV 218
E+Y K+ DLL KKL +++ DG Q+V GL E V + + E+++ GN
Sbjct: 8 EIYGGKLFDLLNDR-----KKLFIRE--DGKQQVCIVGLQEYCVSDVESIKELIERGNAT 166
Query: 219 RSVGSTSANELSSRSH 234
RS G+T ANE SSRSH
Sbjct: 167 RSTGTTGANEESSRSH 214
>AV778230
Length = 463
Score = 37.0 bits (84), Expect = 0.006
Identities = 19/36 (52%), Positives = 21/36 (57%)
Frame = -2
Query: 344 LMFVQISPSSVDLTETLCSLNFATRVRGIESGPARK 379
LMFV ISP E+LCSL FA RV E G R+
Sbjct: 462 LMFVYISPDPSSAGESLCSLRFAARVHACEIGIPRR 355
>AV421105
Length = 415
Score = 35.0 bits (79), Expect = 0.024
Identities = 20/48 (41%), Positives = 29/48 (59%)
Frame = +2
Query: 325 RNSKLTHILQSSLGGDCKTLMFVQISPSSVDLTETLCSLNFATRVRGI 372
R SKLT ILQ SLGG + LM ++P + E++ +++ A R R I
Sbjct: 14 RESKLTRILQDSLGGTSRALMVACLNPG--EYQESVHTVSLAARSRHI 151
>TC10833 similar to UP|Q84V30 (Q84V30) Phosphatidylserine decarboxylase,
partial (37%)
Length = 861
Score = 27.3 bits (59), Expect = 5.0
Identities = 15/28 (53%), Positives = 17/28 (60%), Gaps = 2/28 (7%)
Frame = +1
Query: 199 ETHVYGADGVWEILKSGNRVR--SVGST 224
E VY DGV ILK GN V ++GST
Sbjct: 448 EERVYDCDGVGRILKKGNEVGAFNMGST 531
>TC7807 homologue to UP|Q946J9 (Q946J9) Aquaporin protein PIP1,1, partial
(27%)
Length = 1289
Score = 26.9 bits (58), Expect = 6.5
Identities = 18/39 (46%), Positives = 23/39 (58%)
Frame = +2
Query: 297 LSS*ISLYHHLVMLLQPLLPNQPTYLTDRNSKLTHILQS 335
LSS ISL H L + + P+L N+ T LT TH+L S
Sbjct: 527 LSSLISLDHVLHLTVGPILRNRSTSLT-----FTHMLHS 628
>BP063922
Length = 537
Score = 26.6 bits (57), Expect = 8.5
Identities = 10/32 (31%), Positives = 21/32 (65%)
Frame = +1
Query: 140 LFRVSEERQGTIKYELLVSMLEVYNEKIKDLL 171
++ + + T +Y+ V++ E+YNE+ +DLL
Sbjct: 25 IYLANLDTTSTSQYKFCVTVCELYNEQTRDLL 120
>AV778932
Length = 528
Score = 26.6 bits (57), Expect = 8.5
Identities = 12/35 (34%), Positives = 24/35 (68%)
Frame = -2
Query: 362 SLNFATRVRGIESGPARKQVDLTELLKYKQMAEKS 396
SL+ A R++ I + P+R+++DL + +++ EKS
Sbjct: 233 SLSIAYRLKSIVA*PSRRRLDLKDQMEWYMDKEKS 129
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.326 0.139 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,248,748
Number of Sequences: 28460
Number of extensions: 74540
Number of successful extensions: 513
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 507
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 510
length of query: 430
length of database: 4,897,600
effective HSP length: 93
effective length of query: 337
effective length of database: 2,250,820
effective search space: 758526340
effective search space used: 758526340
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 56 (26.2 bits)
Medicago: description of AC135319.10


