

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135313.7 - phase: 0
(199 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
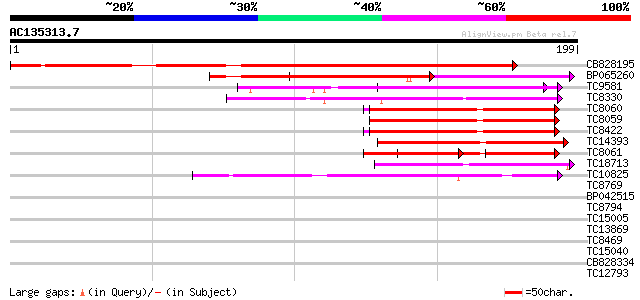
Score E
Sequences producing significant alignments: (bits) Value
CB828195 198 5e-52
BP065260 80 3e-16
TC9581 UP|Q9SCA1 (Q9SCA1) Calcium-binding protein, complete 74 1e-14
TC8330 weakly similar to GB|AAM61257.1|21536925|AY084696 calmodu... 59 7e-10
TC8060 UP|O49184 (O49184) Calmodulin (ESTS AU081349) (Auxin-regu... 54 1e-08
TC8059 GB|AAB68399.1|1773321|HAU79736 calmodulin {Helianthus ann... 54 1e-08
TC8422 UP|AAQ63461 (AAQ63461) Calmodulin 4 (Fragment), complete 54 1e-08
TC14393 similar to UP|Q93YA8 (Q93YA8) Calcium binding protein, p... 54 2e-08
TC8061 UP|AAA34013 (AAA34013) Calmodulin, complete 39 5e-05
TC18713 similar to GB|AAL90989.1|19699246|AY090328 AT3g03000/F13... 42 5e-05
TC10825 similar to GB|AAP68339.1|31711966|BT008900 At1g74740 {Ar... 40 2e-04
TC8769 similar to UP|Q42438 (Q42438) Calcium-dependent protein k... 37 0.002
BP042515 37 0.003
TC8794 similar to UP|O81445 (O81445) Calcineurin B-like protein ... 35 0.009
TC15005 homologue to UP|Q9AR92 (Q9AR92) Protein kinase , partia... 34 0.015
TC13869 weakly similar to UP|Q9FR00 (Q9FR00) Avr9/Cf-9 rapidly e... 33 0.033
TC8469 similar to UP|Q9MAH9 (Q9MAH9) F12M16.12, partial (48%) 33 0.033
TC15040 similar to UP|K22E_HUMAN (P35908) Keratin, type II cytos... 32 0.074
CB828334 32 0.096
TC12793 similar to AAO14864 (AAO14864) Calcineurin B-like protei... 32 0.096
>CB828195
Length = 551
Score = 198 bits (504), Expect = 5e-52
Identities = 108/178 (60%), Positives = 132/178 (73%)
Frame = +3
Query: 1 MALPQISQSNSISNSTSPLFGLIDLFLYLTFFNKIHNFFSSIWFFLLCQIHSGSSEVREE 60
+A Q SQS++ S+S SPLFGLIDLFLY T FNKI FFS+ W +SEVR E
Sbjct: 60 IAKEQTSQSDT-SSSNSPLFGLIDLFLYCTIFNKILKFFSNFW--------CSNSEVRGE 212
Query: 61 KKVSESKCSSQENESNIGRDNGDMIERDEVKMVMEKMGFFCSSESEELEEKYGSKELCEV 120
+KVS+ + + QENES + I+RDEVK VM ++GFFCS ESEELEEKYGSKEL E+
Sbjct: 213 EKVSDYEFNHQENESK-----SEGIQRDEVKKVMAELGFFCSKESEELEEKYGSKELSEL 377
Query: 121 FEENEPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDE 178
FE+ EPSLEE+KQAFDVFDEN+DGFIDA+EL RVL +LGLK+ + E C+ MI FD+
Sbjct: 378 FEDQEPSLEEVKQAFDVFDENRDGFIDARELHRVLCVLGLKEEAGIEKCKIMIRNFDK 551
>BP065260
Length = 446
Score = 79.7 bits (195), Expect = 3e-16
Identities = 42/101 (41%), Positives = 57/101 (55%), Gaps = 1/101 (0%)
Frame = -1
Query: 99 FFCSSESEELEEKYGSKELCEVFEENEPSLEELKQAFDVFD-ENKDGFIDAKELQRVLVI 157
FF + + L++ + P+ E QAFD+F E + + +EL RVL +
Sbjct: 359 FFAAKKVRNLKKSMVPRSFLNCLRTRSPAWRE*XQAFDIFG*EQRWVLLMPRELHRVLCV 180
Query: 158 LGLKQGSEFENCQKMITIFDENQDGRIDFIEFVNIMKNHFC 198
LGLK+ E CQ MI FD+N DGRIDF+EFV IM+N FC
Sbjct: 179 LGLKEEPGIEKCQIMIRNFDKNPDGRIDFLEFVKIMENRFC 57
Score = 78.2 bits (191), Expect = 9e-16
Identities = 44/80 (55%), Positives = 54/80 (67%), Gaps = 1/80 (1%)
Frame = -3
Query: 71 QENESNIGRDNGDMIERDEVKMVMEKMGFFCSSESEELEEKYGSKELCEVFEENEPSLEE 130
QEN+S + I+RDEVK VM ++GFFC ESEELEEKYGS+EL E+FE+ EP LE
Sbjct: 429 QENDSK-----SEGIQRDEVKKVMPELGFFCRQESEELEEKYGSQELSELFEDPEPRLEG 265
Query: 131 LKQAFDVF-DENKDGFIDAK 149
+ F F + GFIDAK
Sbjct: 264 VXPGF*YFWMRTEMGFIDAK 205
>TC9581 UP|Q9SCA1 (Q9SCA1) Calcium-binding protein, complete
Length = 1049
Score = 74.3 bits (181), Expect = 1e-14
Identities = 45/129 (34%), Positives = 72/129 (54%), Gaps = 15/129 (11%)
Frame = +3
Query: 81 NGD-MIERDEVKMVMEKMGFFCSSES-----EELE---------EKYGSKELCEVFEENE 125
NGD I + E+ +E +G F + E ++ +++G EL + +
Sbjct: 381 NGDGRITKKELNDSLENLGIFIPDKELTQMIERIDVNGDGCVDIDEFG--ELYQSIMDER 554
Query: 126 PSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRID 185
E++++AF+VFD+N DGFI +EL+ VL LG+KQG E+C+KMI D + DG +D
Sbjct: 555 DEEEDMREAFNVFDQNGDGFITVEELRTVLASLGIKQGRTVEDCKKMIMKVDVDGDGMVD 734
Query: 186 FIEFVNIMK 194
+ EF +MK
Sbjct: 735 YKEFKQMMK 761
Score = 38.5 bits (88), Expect = 8e-04
Identities = 25/60 (41%), Positives = 31/60 (51%)
Frame = +3
Query: 130 ELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRIDFIEF 189
ELK+ F +FD N DG I KEL L LG+ + +MI D N DG +D EF
Sbjct: 348 ELKRVFQMFDRNGDGRITKKELNDSLENLGIFIPD--KELTQMIERIDVNGDGCVDIDEF 521
>TC8330 weakly similar to GB|AAM61257.1|21536925|AY084696 calmodulin-like
protein {Arabidopsis thaliana;} , partial (72%)
Length = 1174
Score = 58.5 bits (140), Expect = 7e-10
Identities = 41/129 (31%), Positives = 69/129 (52%), Gaps = 11/129 (8%)
Frame = +3
Query: 77 IGRDNGDMIERDEVKMVMEKMGFFCSSESEELE--------EKYGSKELCEVFEENEPSL 128
I RDN ++ R ++ V+ ++G S+ EE+E + GS + + E P
Sbjct: 315 IDRDNDGVVSRADLVAVLTRLGALQLSD-EEVEVMLREVDGDGRGSISVEALMERVGPGS 491
Query: 129 E---ELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRID 185
+ EL++AF+VFD ++DG I A+EL RV +G + + C++MI D N DG +
Sbjct: 492 DPETELREAFEVFDTDRDGRISAEELLRVFTAIG-DERCTLDECRRMIEGVDRNGDGFVC 668
Query: 186 FIEFVNIMK 194
F +F +M+
Sbjct: 669 FEDFSRMME 695
>TC8060 UP|O49184 (O49184) Calmodulin (ESTS AU081349) (Auxin-regulated
calmodulin) (Calmodulin TACAM1-1) (Calmodulin TACAM1-2)
(Calmodulin TACAM1-3) (Calmodulin TACAM3-1) (Calmodulin
TACAM3-2) (Calmodulin TACAM3-3) (Calmodulin TACAM4-1)
(CaM protein), complete
Length = 615
Score = 54.3 bits (129), Expect = 1e-08
Identities = 31/67 (46%), Positives = 44/67 (65%)
Frame = +3
Query: 127 SLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRIDF 186
S EELK+AF VFD++++GFI A EL+ V+ LG K E +MI D + DG+I++
Sbjct: 312 SEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTD--EEVDEMIREADVDGDGQINY 485
Query: 187 IEFVNIM 193
EFV +M
Sbjct: 486 EEFVKVM 506
Score = 48.5 bits (114), Expect = 8e-07
Identities = 28/69 (40%), Positives = 39/69 (55%)
Frame = +3
Query: 125 EPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRI 184
+ + E K+AF +FD++ DG I KEL V+ LG Q Q MI D + +G I
Sbjct: 87 DDQIAEFKEAFSLFDKDGDGCITTKELGTVMRSLG--QNPTEAELQDMINEVDADGNGTI 260
Query: 185 DFIEFVNIM 193
DF EF+N+M
Sbjct: 261 DFPEFLNLM 287
>TC8059 GB|AAB68399.1|1773321|HAU79736 calmodulin {Helianthus annuus;} ,
partial (83%)
Length = 836
Score = 54.3 bits (129), Expect = 1e-08
Identities = 31/67 (46%), Positives = 44/67 (65%)
Frame = +2
Query: 127 SLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRIDF 186
S EELK+AF VFD++++GFI A EL+ V+ LG K E +MI D + DG+I++
Sbjct: 332 SEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTD--EEVDEMIREADVDGDGQINY 505
Query: 187 IEFVNIM 193
EFV +M
Sbjct: 506 EEFVKVM 526
Score = 32.7 bits (73), Expect = 0.043
Identities = 24/56 (42%), Positives = 29/56 (50%), Gaps = 1/56 (1%)
Frame = +2
Query: 139 DENKD-GFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRIDFIEFVNIM 193
DE D G I KEL V+ LG Q Q MI D + +G IDF EF+N+M
Sbjct: 146 DEAADPGCITTKELGTVMRSLG--QNPTEAELQDMINEVDADGNGTIDFPEFLNLM 307
>TC8422 UP|AAQ63461 (AAQ63461) Calmodulin 4 (Fragment), complete
Length = 832
Score = 54.3 bits (129), Expect = 1e-08
Identities = 31/67 (46%), Positives = 44/67 (65%)
Frame = +1
Query: 127 SLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRIDF 186
S EELK+AF VFD++++GFI A EL+ V+ LG K E +MI D + DG+I++
Sbjct: 379 SEEELKEAFRVFDKDQNGFISAAELRHVMTNLGEKLTD--EEVDEMIREADVDGDGQINY 552
Query: 187 IEFVNIM 193
EFV +M
Sbjct: 553 EEFVKVM 573
Score = 48.9 bits (115), Expect = 6e-07
Identities = 28/69 (40%), Positives = 39/69 (55%)
Frame = +1
Query: 125 EPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRI 184
+ + E K+AF +FD++ DG I KEL V+ LG Q Q MI D + +G I
Sbjct: 154 DDQISEFKEAFSLFDKDGDGSITTKELGTVMRSLG--QNPTEAELQDMINEVDADGNGTI 327
Query: 185 DFIEFVNIM 193
DF EF+N+M
Sbjct: 328 DFPEFLNLM 354
>TC14393 similar to UP|Q93YA8 (Q93YA8) Calcium binding protein, partial
(85%)
Length = 786
Score = 53.5 bits (127), Expect = 2e-08
Identities = 28/67 (41%), Positives = 41/67 (60%)
Frame = +3
Query: 130 ELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRIDFIEF 189
EL AF+++D++K+G I A EL +VL LG+ E C KMI D + DG ++F EF
Sbjct: 366 ELHDAFELYDQDKNGLISAAELCKVLNRLGMNCSEE--ECHKMIKSVDSDGDGNVNFEEF 539
Query: 190 VNIMKNH 196
+M N+
Sbjct: 540 KKMMTNN 560
>TC8061 UP|AAA34013 (AAA34013) Calmodulin, complete
Length = 568
Score = 39.3 bits (90), Expect = 5e-04
Identities = 22/57 (38%), Positives = 35/57 (60%)
Frame = +2
Query: 137 VFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRIDFIEFVNIM 193
+ D++++GFI A EL+ V+ LG K E +MI D + DG+I++ EFV +M
Sbjct: 320 LLDKDQNGFISAAELRHVMTNLGEKLTDE--EVDEMIREADVDGDGQINYEEFVKVM 484
Score = 32.7 bits (73), Expect(2) = 5e-05
Identities = 15/35 (42%), Positives = 22/35 (62%)
Frame = +3
Query: 125 EPSLEELKQAFDVFDENKDGFIDAKELQRVLVILG 159
+ + E K+AF +FD++ DG I KEL V+ LG
Sbjct: 63 DEQISEFKEAFSLFDKDGDGCITTKELGTVMRSLG 167
Score = 28.9 bits (63), Expect(2) = 5e-05
Identities = 12/26 (46%), Positives = 18/26 (69%)
Frame = +1
Query: 168 NCQKMITIFDENQDGRIDFIEFVNIM 193
+C+ MI D + +G IDF EF+N+M
Sbjct: 187 SCRHMINEVDADGNGTIDFPEFLNLM 264
>TC18713 similar to GB|AAL90989.1|19699246|AY090328 AT3g03000/F13E7_5
{Arabidopsis thaliana;}, partial (88%)
Length = 868
Score = 42.4 bits (98), Expect = 5e-05
Identities = 29/81 (35%), Positives = 37/81 (44%), Gaps = 11/81 (13%)
Frame = +1
Query: 129 EELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRIDFIE 188
E+L+Q F VFD + +GFI A EL + LG E MI D + DG I F E
Sbjct: 439 EQLRQLFRVFDRDGNGFITAAELAHSMAKLG--HALSAEELTGMIKEADADGDGMISFQE 612
Query: 189 FVNIMK-----------NHFC 198
F + + NHFC
Sbjct: 613 FAHAISSAAFDNSWA*LNHFC 675
>TC10825 similar to GB|AAP68339.1|31711966|BT008900 At1g74740 {Arabidopsis
thaliana;}, partial (32%)
Length = 1091
Score = 40.4 bits (93), Expect = 2e-04
Identities = 33/131 (25%), Positives = 61/131 (46%), Gaps = 1/131 (0%)
Frame = +1
Query: 65 ESKCSSQENESNIGRDNGDMIERDEVKMVMEKMGFFCSSESEELEEKYGSKELCEVFEEN 124
+ + S +E ++ + R G + E+KM+ME + + YG + +
Sbjct: 55 DGRVSYEELKAGL-RKVGSQLADQEIKMLMEVADVDGNGVLD-----YGEFVAVTIHLQK 216
Query: 125 EPSLEELKQAFDVFDENKDGFIDAKELQRVLV-ILGLKQGSEFENCQKMITIFDENQDGR 183
+ E +AF FD++ G+I++ ELQ L G+ + + + D ++DGR
Sbjct: 217 MENDEHFHKAFKFFDKDGSGYIESGELQEALADESGVTDADVLNDIMREV---DTDKDGR 387
Query: 184 IDFIEFVNIMK 194
I + EFV +MK
Sbjct: 388 ISYEEFVAMMK 420
>TC8769 similar to UP|Q42438 (Q42438) Calcium-dependent protein kinase ,
partial (21%)
Length = 1144
Score = 37.0 bits (84), Expect = 0.002
Identities = 24/71 (33%), Positives = 39/71 (54%), Gaps = 5/71 (7%)
Frame = +2
Query: 129 EELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFE-NCQKMITIF----DENQDGR 183
E L +AF FD N+ G+I+ +EL+ L E E N +++I+ D ++DG+
Sbjct: 62 EHLIKAFKFFDANQSGYIEIEELRDAL-------SDEVETNSEEVISAIMHDVDTDKDGK 220
Query: 184 IDFIEFVNIMK 194
I + EF +MK
Sbjct: 221 ISYEEFATMMK 253
>BP042515
Length = 539
Score = 36.6 bits (83), Expect = 0.003
Identities = 21/57 (36%), Positives = 32/57 (55%)
Frame = -1
Query: 138 FDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRIDFIEFVNIMK 194
FD++ G+I ELQ+ GL ++MI D++ DGRID+ EFV +M+
Sbjct: 539 FDKDGSGYITQDELQQACEEFGLGD----VRLEEMIREADQDNDGRIDYNEFVAMMQ 381
>TC8794 similar to UP|O81445 (O81445) Calcineurin B-like protein 1,
complete
Length = 1077
Score = 35.0 bits (79), Expect = 0.009
Identities = 20/76 (26%), Positives = 40/76 (52%), Gaps = 6/76 (7%)
Frame = +3
Query: 120 VFEENEPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFEN------CQKMI 173
VF N +++ +F ++D + GFI+ +E++++L+ L + + + K
Sbjct: 471 VFHPNAALEDKIDFSFRLYDLDSTGFIERQEVKQMLIALLCESEMKLADEVVETIIDKTF 650
Query: 174 TIFDENQDGRIDFIEF 189
D+NQDG+ID E+
Sbjct: 651 VDADQNQDGKIDIDEW 698
Score = 26.9 bits (58), Expect = 2.4
Identities = 17/68 (25%), Positives = 35/68 (51%), Gaps = 4/68 (5%)
Frame = +3
Query: 127 SLEELKQAFDVFDENKDGFID----AKELQRVLVILGLKQGSEFENCQKMITIFDENQDG 182
++ E++ F++F +D +KE ++ + K+ + F N ++ +FD + G
Sbjct: 261 TVSEVEALFELFKSISSSVVDDGLISKEEFQLAIFKNRKKENIFAN--RIFDLFDVKKKG 434
Query: 183 RIDFIEFV 190
IDF +FV
Sbjct: 435 VIDFDDFV 458
>TC15005 homologue to UP|Q9AR92 (Q9AR92) Protein kinase , partial (19%)
Length = 657
Score = 34.3 bits (77), Expect = 0.015
Identities = 20/71 (28%), Positives = 36/71 (50%), Gaps = 4/71 (5%)
Frame = +2
Query: 129 EELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFE----NCQKMITIFDENQDGRI 184
+ L AF FD++ GFI EL+ + G+ + + +I+ D + DGRI
Sbjct: 80 DHLN*AFPYFDKDSSGFITRDELEIAMKEYGMGDDATIKEIISEVDTIISEVDTDHDGRI 259
Query: 185 DFIEFVNIMKN 195
++ EF +M++
Sbjct: 260 NYEEFCAMMRS 292
>TC13869 weakly similar to UP|Q9FR00 (Q9FR00) Avr9/Cf-9 rapidly elicited
protein 31, partial (45%)
Length = 547
Score = 33.1 bits (74), Expect = 0.033
Identities = 17/43 (39%), Positives = 26/43 (59%), Gaps = 2/43 (4%)
Frame = +3
Query: 121 FEENEPSLEELK--QAFDVFDENKDGFIDAKELQRVLVILGLK 161
F PSL L+ + FD+FD+N D I +E+ + L +LGL+
Sbjct: 252 FRLRSPSLNSLRLRRIFDMFDKNGDCMITVEEISQALNLLGLE 380
>TC8469 similar to UP|Q9MAH9 (Q9MAH9) F12M16.12, partial (48%)
Length = 1039
Score = 33.1 bits (74), Expect = 0.033
Identities = 14/35 (40%), Positives = 22/35 (62%)
Frame = +2
Query: 122 EENEPSLEELKQAFDVFDENKDGFIDAKELQRVLV 156
E+ +P E +K+ F DENKDG + EL+ ++V
Sbjct: 863 EDGQPDTEVIKKLFTTIDENKDGSLTHGELRALVV 967
>TC15040 similar to UP|K22E_HUMAN (P35908) Keratin, type II cytoskeletal 2
epidermal (Cytokeratin 2e) (K2e) (CK 2e), partial (3%)
Length = 573
Score = 32.0 bits (71), Expect = 0.074
Identities = 14/26 (53%), Positives = 20/26 (76%)
Frame = +3
Query: 130 ELKQAFDVFDENKDGFIDAKELQRVL 155
E+ +AF + D ++ GFID KELQ+VL
Sbjct: 354 EVIRAFQMVDRDQSGFIDEKELQQVL 431
>CB828334
Length = 541
Score = 31.6 bits (70), Expect = 0.096
Identities = 17/66 (25%), Positives = 31/66 (46%)
Frame = +2
Query: 125 EPSLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENCQKMITIFDENQDGRI 184
E + E L F D+NKDG++ E+ + + + S K D +++G +
Sbjct: 308 ETTFETLVDTFVFLDKNKDGYVSKSEMVQAINETVSGERSSGRIAMKRFEEMDWDKNGMV 487
Query: 185 DFIEFV 190
+F EF+
Sbjct: 488 NFKEFL 505
>TC12793 similar to AAO14864 (AAO14864) Calcineurin B-like protein, partial
(44%)
Length = 534
Score = 31.6 bits (70), Expect = 0.096
Identities = 19/74 (25%), Positives = 41/74 (54%), Gaps = 3/74 (4%)
Frame = +1
Query: 123 ENEP-SLEELKQAFDVFDENKDGFIDAKELQRVLVILGLKQGSEFENC--QKMITIFDEN 179
+N P ++ E++ +++F + ID + + + L L + S +N ++ +FDE
Sbjct: 211 KNSPFTVNEIEALYELFKKLSSSVIDDGLIHKEELTLALLKTSTGKNLFLDRVFDLFDEK 390
Query: 180 QDGRIDFIEFVNIM 193
++G I+F EFV+ +
Sbjct: 391 RNGVIEFEEFVHAL 432
Score = 25.8 bits (55), Expect = 5.3
Identities = 10/27 (37%), Positives = 17/27 (62%)
Frame = +1
Query: 131 LKQAFDVFDENKDGFIDAKELQRVLVI 157
L + FD+FDE ++G I+ +E L +
Sbjct: 358 LDRVFDLFDEKRNGVIEFEEFVHALSV 438
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.136 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,374,380
Number of Sequences: 28460
Number of extensions: 44101
Number of successful extensions: 424
Number of sequences better than 10.0: 67
Number of HSP's better than 10.0 without gapping: 399
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 412
length of query: 199
length of database: 4,897,600
effective HSP length: 86
effective length of query: 113
effective length of database: 2,450,040
effective search space: 276854520
effective search space used: 276854520
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 53 (25.0 bits)
Medicago: description of AC135313.7


