

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135230.5 + phase: 0
(412 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
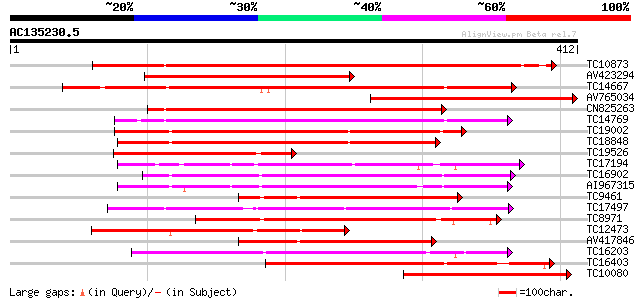
Score E
Sequences producing significant alignments: (bits) Value
TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partia... 416 e-117
AV423294 281 2e-76
TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, com... 249 8e-67
AV765034 238 1e-63
CN825263 231 2e-61
TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete 225 9e-60
TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866)... 221 2e-58
TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, pa... 207 2e-54
TC19526 homologue to UP|AAQ96340 (AAQ96340) Protein kinase-like ... 205 1e-53
TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete 202 8e-53
TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-... 200 3e-52
AI967315 199 7e-52
TC9461 similar to UP|O49840 (O49840) Protein kinase (AT2G02800/T... 197 3e-51
TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2,... 192 1e-49
TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P... 185 1e-47
TC12473 similar to UP|Q9M068 (Q9M068) Protein kinase-like protei... 177 3e-45
AV417846 177 4e-45
TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodula... 176 6e-45
TC16403 homologue to UP|Q9LKY4 (Q9LKY4) Pti1 kinase-like protein... 174 3e-44
TC10080 similar to UP|AAQ96340 (AAQ96340) Protein kinase-like pr... 169 1e-42
>TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partial (77%)
Length = 1478
Score = 416 bits (1069), Expect = e-117
Identities = 206/337 (61%), Positives = 260/337 (77%)
Frame = +1
Query: 61 KNLNLNDGVSPDGKVAQTFTFAELAAATENFRADCFVGEGGFGKVYKGYLEKINQVVAIK 120
K ++ ++G S A +F F ELA AT NF+ +GEGGFGKVYKG L + VA+K
Sbjct: 361 KGVSSSNGSSNGKTAAASFGFRELADATRNFKEANLIGEGGFGKVYKGRLTT-GEAVAVK 537
Query: 121 QLDRNGVQGIREFVVEVITLGLADHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDL 180
QL +G QG +EFV+EV+ L L H NLV+L+G+C +G+QRLLVYEYMP+GSLE+HL +L
Sbjct: 538 QLSHDGRQGFQEFVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFEL 717
Query: 181 SPGQKPLDWNTRMKIAAGAARGLEYLHDKMKPPVIYRDLKCSNILLGEDYHSKLSDFGLA 240
S ++PL+W+TRMK+A GAARGLEYLH PPVIYRDLK +NILL +++ KLSDFGLA
Sbjct: 718 SHDKEPLNWSTRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLA 897
Query: 241 KVGPIGDKTHVSTRVMGTYGYCAPDYAMTGQLTFKSDIYSFGVALLELITGRKAIDHKKP 300
K+GP+GD THVSTRVMGTYGYCAP+YAM+G+LT KSDIYSFGV LLEL+TGR+AID +
Sbjct: 898 KLGPVGDNTHVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRR 1077
Query: 301 AKEQNLVAWARPLFRDRRRFSEMIDPLLEGQYPVRGLYQALAIAAMCVQEQPNMRPVIAD 360
EQNLV+WARP F DRRRF M+DPLL+G++P R L+QA+AI AMC+QEQP RP+I D
Sbjct: 1078PGEQNLVSWARPYFSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITD 1257
Query: 361 VVTALNYLASQKYDPQIHHIQGSRKGSSSPRSRSERH 397
+V AL YLASQ P+ H++ G ++ P SR ER+
Sbjct: 1258IVVALEYLASQG-SPEAHYLYGVQQ----PPSRMERN 1353
>AV423294
Length = 459
Score = 281 bits (718), Expect = 2e-76
Identities = 130/152 (85%), Positives = 147/152 (96%)
Frame = +3
Query: 99 EGGFGKVYKGYLEKINQVVAIKQLDRNGVQGIREFVVEVITLGLADHPNLVKLLGFCAEG 158
EGGFGKVY+G++E+INQVVAIKQL+ +G+QGIREFVVEV+TL LADHPNLVKL+GFCAEG
Sbjct: 3 EGGFGKVYRGFIERINQVVAIKQLNPHGLQGIREFVVEVLTLSLADHPNLVKLIGFCAEG 182
Query: 159 EQRLLVYEYMPLGSLENHLHDLSPGQKPLDWNTRMKIAAGAARGLEYLHDKMKPPVIYRD 218
EQRLLVYEYMPLGSLE+HLHD+ PG+KPLDWNTRMKIAAGAARGLEYLH+KMKPPVIYRD
Sbjct: 183 EQRLLVYEYMPLGSLESHLHDILPGKKPLDWNTRMKIAAGAARGLEYLHNKMKPPVIYRD 362
Query: 219 LKCSNILLGEDYHSKLSDFGLAKVGPIGDKTH 250
+KCSN+L+G+ YH KLSDFGLAKVGPIGDKTH
Sbjct: 363 IKCSNLLIGDGYHPKLSDFGLAKVGPIGDKTH 458
>TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, complete
Length = 1591
Score = 249 bits (635), Expect = 8e-67
Identities = 140/336 (41%), Positives = 206/336 (60%), Gaps = 6/336 (1%)
Frame = +2
Query: 39 NLSELKDKKEDDSKHDQLSLDVKNLNLNDGVSPDGKVAQTFTFAELAAATENFRADCFVG 98
+L ++ + +SK + S VK+ +P + EL T+NF + +G
Sbjct: 212 HLKSPRNYGDGNSKGSKASAPVKHETQK---APPPIEVPALSLDELKEKTDNFGSKALIG 382
Query: 99 EGGFGKVYKGYLEKINQVVAIKQLDRNGV-QGIREFVVEVITLGLADHPNLVKLLGFCAE 157
EG +G+VY L N VA+K+LD + + EF+ +V + + N V+L G+C E
Sbjct: 383 EGSYGRVYYATLNDGN-AVAVKKLDVSSEPETNNEFLTQVSMVSRLKNDNFVELHGYCVE 559
Query: 158 GEQRLLVYEYMPLGSLENHLHDLS--PGQKP---LDWNTRMKIAAGAARGLEYLHDKMKP 212
G R+L YE+ +GSL + LH G +P LDW R++IA AARGLEYLH+K++P
Sbjct: 560 GNLRVLAYEFATMGSLHDILHGRKGVQGAQPGPTLDWIQRVRIAVDAARGLEYLHEKVQP 739
Query: 213 PVIYRDLKCSNILLGEDYHSKLSDFGLAKVGPIGDKTHVSTRVMGTYGYCAPDYAMTGQL 272
+I+RD++ SN+L+ EDY +K++DF L+ P STRV+GT+GY AP+YAMTGQL
Sbjct: 740 AIIHRDIRSSNVLIFEDYKAKIADFNLSNQAPDMAARLHSTRVLGTFGYHAPEYAMTGQL 919
Query: 273 TFKSDIYSFGVALLELITGRKAIDHKKPAKEQNLVAWARPLFRDRRRFSEMIDPLLEGQY 332
T KSD+YSFGV LLEL+TGRK +DH P +Q+LV WA P + + + +DP L+G+Y
Sbjct: 920 TQKSDVYSFGVVLLELLTGRKPVDHTMPRGQQSLVTWATPRLSE-DKVKQCVDPKLKGEY 1096
Query: 333 PVRGLYQALAIAAMCVQEQPNMRPVIADVVTALNYL 368
P +G+ + A+AA+CVQ + RP ++ VV AL L
Sbjct: 1097PPKGVAKLAAVAALCVQYEAEFRPNMSIVVKALQPL 1204
>AV765034
Length = 529
Score = 238 bits (607), Expect = 1e-63
Identities = 118/151 (78%), Positives = 130/151 (85%), Gaps = 1/151 (0%)
Frame = -3
Query: 263 APDYAMTGQLTFKSDIYSFGVALLELITGRKAIDHKKPAKEQNLVAWARPLFRDRRRFSE 322
APDYAMTGQLTFKSD+YSFGV LLE+ITGRKAIDH KPAKEQNLVAWARPLFRDR++FS
Sbjct: 527 APDYAMTGQLTFKSDVYSFGVVLLEIITGRKAIDHTKPAKEQNLVAWARPLFRDRKKFSL 348
Query: 323 MIDPLLEGQYPVRGLYQALAIAAMCVQEQPNMRPVIADVVTALNYLASQKYDPQIHHIQG 382
M+DPLLEGQYPVRGLYQALAIAAMCVQEQPNMRPVI DVVTALNYLA+ KYDPQ ++
Sbjct: 347 MVDPLLEGQYPVRGLYQALAIAAMCVQEQPNMRPVIVDVVTALNYLATNKYDPQNQTVES 168
Query: 383 S-RKGSSSPRSRSERHRRVTSKDSETDRFGD 412
S R+ +SSP + + HRR S DS TD GD
Sbjct: 167 SNRRNTSSPMGKGDDHRRNDSNDSGTDISGD 75
>CN825263
Length = 663
Score = 231 bits (588), Expect = 2e-61
Identities = 118/217 (54%), Positives = 146/217 (66%)
Frame = +1
Query: 101 GFGKVYKGYLEKINQVVAIKQLDRNGVQGIREFVVEVITLGLADHPNLVKLLGFCAEGEQ 160
GFG VYKG L + VA+K L R+ +G REF+ EV L H NLVKL+G C E +
Sbjct: 1 GFGLVYKGILND-GRDVAVKILKRDDQRGGREFLAEVEMLSRLHHRNLVKLIGICIEKQT 177
Query: 161 RLLVYEYMPLGSLENHLHDLSPGQKPLDWNTRMKIAAGAARGLEYLHDKMKPPVIYRDLK 220
R L+YE +P GS+E+HLH PLDWN RMKIA GAARGL YLH+ P VI+RD K
Sbjct: 178 RCLIYELVPNGSVESHLHGADKETGPLDWNARMKIALGAARGLAYLHEDSNPCVIHRDFK 357
Query: 221 CSNILLGEDYHSKLSDFGLAKVGPIGDKTHVSTRVMGTYGYCAPDYAMTGQLTFKSDIYS 280
SNILL D+ K+SDFGLA+ H+ST VMGT+GY AP+YAMTG L KSD+YS
Sbjct: 358 SSNILLECDFTPKVSDFGLARTALDEGNKHISTHVMGTFGYLAPEYAMTGHLLVKSDVYS 537
Query: 281 FGVALLELITGRKAIDHKKPAKEQNLVAWARPLFRDR 317
+GV LLEL+TG K +D +P ++NLV WARP+ +
Sbjct: 538 YGVVLLELLTGTKPVDLSQPPGQENLVTWARPILTSK 648
>TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete
Length = 3495
Score = 225 bits (574), Expect = 9e-60
Identities = 122/289 (42%), Positives = 173/289 (59%)
Frame = +3
Query: 77 QTFTFAELAAATENFRADCFVGEGGFGKVYKGYLEKINQVVAIKQLDRNGVQGIREFVVE 136
Q FT + ATE ++ +GEGGFG VY+G L Q VA+K QG REF E
Sbjct: 2127 QAFTLEYIEVATERYKT--LIGEGGFGSVYRGTLND-GQEVAVKVRSSTSTQGTREFDNE 2297
Query: 137 VITLGLADHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLSPGQKPLDWNTRMKIA 196
+ L H NLV LLG+C E +Q++LVY +M GSL++ L+ +K LDW TR+ IA
Sbjct: 2298 LNLLSAIQHENLVPLLGYCNESDQQILVYPFMSNGSLQDRLYGEPAKRKILDWPTRLSIA 2477
Query: 197 AGAARGLEYLHDKMKPPVIYRDLKCSNILLGEDYHSKLSDFGLAKVGPIGDKTHVSTRVM 256
GAARGL YLH VI+RD+K SNILL +K++DFG +K P ++VS V
Sbjct: 2478 LGAARGLAYLHTFPGRSVIHRDIKSSNILLDHSMCAKVADFGFSKYAPQEGDSYVSLEVR 2657
Query: 257 GTYGYCAPDYAMTGQLTFKSDIYSFGVALLELITGRKAIDHKKPAKEQNLVAWARPLFRD 316
GT GY P+Y T QL+ KSD++SFGV LLE+++GR+ ++ K+P E +LV WA P R
Sbjct: 2658 GTAGYLDPEYYKTQQLSEKSDVFSFGVVLLEIVSGREPLNIKRPRTEWSLVEWATPYIRG 2837
Query: 317 RRRFSEMIDPLLEGQYPVRGLYQALAIAAMCVQEQPNMRPVIADVVTAL 365
+ E++DP ++G Y +++ + +A C++ RP + +V L
Sbjct: 2838 -SKVDEIVDPGIKGGYHAEAMWRVVEVALQCLEPFSTYRPSMVAIVREL 2981
>TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866), partial
(48%)
Length = 780
Score = 221 bits (562), Expect = 2e-58
Identities = 117/256 (45%), Positives = 165/256 (63%)
Frame = +1
Query: 77 QTFTFAELAAATENFRADCFVGEGGFGKVYKGYLEKINQVVAIKQLDRNGVQGIREFVVE 136
+ F+ EL +AT NF D +GEGGFG VY G L +Q+ A+K+L + EF VE
Sbjct: 22 RVFSLKELHSATNNFNYDNKLGEGGFGSVYWGQLWDGSQI-AVKRLKVWSNKADMEFAVE 198
Query: 137 VITLGLADHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLSPGQKPLDWNTRMKIA 196
V L H NL+ L G+CAEG++RL+VY+YMP SL +HLH + LDWN RM IA
Sbjct: 199 VEILARVRHKNLLSLRGYCAEGQERLIVYDYMPNLSLLSHLHGQHSSECLLDWNRRMNIA 378
Query: 197 AGAARGLEYLHDKMKPPVIYRDLKCSNILLGEDYHSKLSDFGLAKVGPIGDKTHVSTRVM 256
G+A G+ YLH + P +I+RD+K SN+LL D+ ++++DFG AK+ P G THV+TRV
Sbjct: 379 IGSAEGIVYLHHQATPHIIHRDIKASNVLLDSDFQARVADFGFAKLIPDG-ATHVTTRVK 555
Query: 257 GTYGYCAPDYAMTGQLTFKSDIYSFGVALLELITGRKAIDHKKPAKEQNLVAWARPLFRD 316
GT GY AP+YAM G+ D++SFG+ LLEL +G+K ++ ++++ WA PL
Sbjct: 556 GTLGYLAPEYAMLGKANECCDVFSFGILLLELASGKKPLEKLSSTVKRSINDWALPL-AC 732
Query: 317 RRRFSEMIDPLLEGQY 332
++F+E DP L G+Y
Sbjct: 733 AKKFTEFADPRLNGEY 780
>TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, partial (64%)
Length = 969
Score = 207 bits (528), Expect = 2e-54
Identities = 110/235 (46%), Positives = 150/235 (63%)
Frame = +2
Query: 79 FTFAELAAATENFRADCFVGEGGFGKVYKGYLEKINQVVAIKQLDRNGVQGIREFVVEVI 138
FT+ EL AAT F D +GEGGFG VY G Q+ A+K+L + EF VEV
Sbjct: 266 FTYKELHAATGGFSDDNKLGEGGFGSVYWGRTSDGLQI-AVKKLKAMNSKAEMEFAVEVE 442
Query: 139 TLGLADHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLSPGQKPLDWNTRMKIAAG 198
LG H NL+ L G+C +QRL+VY+YMP SL +HLH + L+W RMKIA G
Sbjct: 443 VLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQFAVEVQLNWQKRMKIAIG 622
Query: 199 AARGLEYLHDKMKPPVIYRDLKCSNILLGEDYHSKLSDFGLAKVGPIGDKTHVSTRVMGT 258
+A G+ YLH ++ P +I+RD+K SN+LL D+ ++DFG AK+ P G +H++TRV GT
Sbjct: 623 SAEGILYLHHEVTPHIIHRDIKASNVLLNSDFEPLVADFGFAKLIPEG-VSHMTTRVKGT 799
Query: 259 YGYCAPDYAMTGQLTFKSDIYSFGVALLELITGRKAIDHKKPAKEQNLVAWARPL 313
GY AP+YAM G+++ D+YSFG+ LLEL+TGRK I+ ++ + WA PL
Sbjct: 800 LGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGVKRTITEWAEPL 964
>TC19526 homologue to UP|AAQ96340 (AAQ96340) Protein kinase-like protein,
partial (37%)
Length = 432
Score = 205 bits (521), Expect = 1e-53
Identities = 98/133 (73%), Positives = 115/133 (85%)
Frame = +1
Query: 76 AQTFTFAELAAATENFRADCFVGEGGFGKVYKGYLEKINQVVAIKQLDRNGVQGIREFVV 135
AQTFTF ELAAAT+NFR +C +GEGGFG+VYKG LE QVVA+KQLDRNG+QG REF+V
Sbjct: 43 AQTFTFRELAAATKNFRPECLLGEGGFGRVYKGRLESTGQVVAVKQLDRNGLQGNREFLV 222
Query: 136 EVITLGLADHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLSPGQKPLDWNTRMKI 195
EV+ L L HPNLV L+G+CA+G+QRLLVYE+MPLGSLE+HLH ++PLDWNTRMKI
Sbjct: 223 EVLMLSLLHHPNLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHG---DKEPLDWNTRMKI 393
Query: 196 AAGAARGLEYLHD 208
AAGAA+GLEYLHD
Sbjct: 394 AAGAAKGLEYLHD 432
>TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete
Length = 2193
Score = 202 bits (514), Expect = 8e-53
Identities = 119/305 (39%), Positives = 183/305 (59%), Gaps = 9/305 (2%)
Frame = +1
Query: 79 FTFAELAAATENFRADCFVGEGGFGKVYKGYLEKINQVVAIKQLDRNGVQGIREFVVEVI 138
F++ ELA AT NF D +G+GGFG VY Y E + AIK++D VQ EF+ E+
Sbjct: 1051 FSYQELAKATNNFSLDNKIGQGGFGAVY--YAELRGKKTAIKKMD---VQASTEFLCELK 1215
Query: 139 TLGLADHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLSPGQKPLDWNTRMKIAAG 198
L H NLV+L+G+C EG LVYE++ G+L +LH G++PL W++R++IA
Sbjct: 1216 VLTHVHHLNLVRLIGYCVEGSL-FLVYEHIDNGNLGQYLH--GSGKEPLPWSSRVQIALD 1386
Query: 199 AARGLEYLHDKMKPPVIYRDLKCSNILLGEDYHSKLSDFGLAKVGPIGDKTHVSTRVMGT 258
AARGLEY+H+ P I+RD+K +NIL+ ++ K++DFGL K+ +G+ T + TR++GT
Sbjct: 1387 AARGLEYIHEHTVPVYIHRDVKSANILIDKNLRGKVADFGLTKLIEVGNST-LQTRLVGT 1563
Query: 259 YGYCAPDYAMTGQLTFKSDIYSFGVALLELITGRKAI--DHKKPAKEQNLVAWARPLFRD 316
+GY P+YA G ++ K D+Y+FGV L ELI+ + A+ + A+ + LVA LF +
Sbjct: 1564 FGYMPPEYAQYGDISPKIDVYAFGVVLFELISAKNAVLKTGELVAESKGLVA----LFEE 1731
Query: 317 RRRFSE-------MIDPLLEGQYPVRGLYQALAIAAMCVQEQPNMRPVIADVVTALNYLA 369
S+ ++DP L YP+ + + + C ++ P +RP + +V AL L+
Sbjct: 1732 ALNKSDPCDALRKLVDPRLGENYPIDSVLKIAQLGRACTRDNPLLRPSMRSLVVALMTLS 1911
Query: 370 SQKYD 374
S D
Sbjct: 1912 SLTED 1926
>TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-like
protein, partial (46%)
Length = 941
Score = 200 bits (509), Expect = 3e-52
Identities = 110/271 (40%), Positives = 162/271 (59%)
Frame = +3
Query: 97 VGEGGFGKVYKGYLEKINQVVAIKQLDRNGVQGIREFVVEVITLGLADHPNLVKLLGFCA 156
+G GGFGKVY G ++ +V AIK+ + QG+ EF E+ L H +LV L+G+C
Sbjct: 15 LGVGGFGKVYYGEVDGGTKV-AIKRGNPLSEQGVHEFQTEIEMLSKLRHRHLVSLIGYCE 191
Query: 157 EGEQRLLVYEYMPLGSLENHLHDLSPGQKPLDWNTRMKIAAGAARGLEYLHDKMKPPVIY 216
E + +LVY++M G+L HL+ + PL W R++I GAARGL YLH K +I+
Sbjct: 192 ENTEMILVYDHMAYGTLREHLYKTQ--KPPLPWKQRLEICIGAARGLHYLHTGAKYTIIH 365
Query: 217 RDLKCSNILLGEDYHSKLSDFGLAKVGPIGDKTHVSTRVMGTYGYCAPDYAMTGQLTFKS 276
RD+K +NILL E + +K+SDFGL+K GP D THVST V G++GY P+Y QLT KS
Sbjct: 366 RDVKTTNILLDEKWVAKVSDFGLSKTGPTLDNTHVSTVVKGSFGYLDPEYFRRQQLTDKS 545
Query: 277 DIYSFGVALLELITGRKAIDHKKPAKEQNLVAWARPLFRDRRRFSEMIDPLLEGQYPVRG 336
D+YSFGV L E++ R A++ ++ +L WA + ++ +++DP L+G+
Sbjct: 546 DVYSFGVVLFEILCARPALNPSLAKEQVSLAEWASHCY-NKGILDQILDPYLKGKIAPEC 722
Query: 337 LYQALAIAAMCVQEQPNMRPVIADVVTALNY 367
+ A CV +Q RP + DV+ L +
Sbjct: 723 FKKFAETAMKCVSDQGIERPSMGDVLWNLEF 815
>AI967315
Length = 1308
Score = 199 bits (506), Expect = 7e-52
Identities = 112/289 (38%), Positives = 173/289 (59%), Gaps = 2/289 (0%)
Frame = +1
Query: 79 FTFAELAAATENFRADCFVGEGGFGKVYKGYLEKINQVVAIKQLDRN--GVQGIREFVVE 136
F++ EL AT F ++ VG+GG+ +VYKG LE +++ A+K+L R + +EF+ E
Sbjct: 112 FSYEELFHATNGFSSENMVGKGGYAEVYKGRLESGDEI-AVKRLTRTCRDERKEKEFLTE 288
Query: 137 VITLGLADHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLSPGQKPLDWNTRMKIA 196
+ T+G H N++ LLG C + LV+E +GS+ + +HD PLDW TR KI
Sbjct: 289 IGTIGHVCHSNVMPLLGCCIDNGL-YLVFELSTVGSVASLIHDEK--MAPLDWKTRYKIV 459
Query: 197 AGAARGLEYLHDKMKPPVIYRDLKCSNILLGEDYHSKLSDFGLAKVGPIGDKTHVSTRVM 256
G ARGL YLH + +I+RD+K SNILL ED+ ++SDFGLAK P H +
Sbjct: 460 LGTARGLHYLHKGCQRRIIHRDIKASNILLTEDFEPQISDFGLAKWLPSQWTHHSIAPIE 639
Query: 257 GTYGYCAPDYAMTGQLTFKSDIYSFGVALLELITGRKAIDHKKPAKEQNLVAWARPLFRD 316
GT+G+ AP+Y M G + K+D+++FGV LLE+I+GRK +D Q+L WA+P+
Sbjct: 640 GTFGHLAPEYYMHGVVDEKTDVFAFGVFLLEVISGRKPVD----GSHQSLHTWAKPIL-S 804
Query: 317 RRRFSEMIDPLLEGQYPVRGLYQALAIAAMCVQEQPNMRPVIADVVTAL 365
+ +++DP LEG Y V + A++C++ RP +++V+ +
Sbjct: 805 KWEIEKLVDPRLEGCYDVTQFNRVAFAASLCIRASSTWRPTMSEVLEVM 951
>TC9461 similar to UP|O49840 (O49840) Protein kinase (AT2G02800/T20F6.6) ,
partial (37%)
Length = 484
Score = 197 bits (500), Expect = 3e-51
Identities = 98/163 (60%), Positives = 121/163 (74%)
Frame = +3
Query: 167 YMPLGSLENHLHDLSPGQKPLDWNTRMKIAAGAARGLEYLHDKMKPPVIYRDLKCSNILL 226
+MP GSLENHL P +PL W+ RMK+A GAARGL +LH+ K VIYRD K SNILL
Sbjct: 3 FMPKGSLENHLFRRGP--QPLSWSVRMKVAIGAARGLSFLHNA-KSQVIYRDFKASNILL 173
Query: 227 GEDYHSKLSDFGLAKVGPIGDKTHVSTRVMGTYGYCAPDYAMTGQLTFKSDIYSFGVALL 286
++++KLSDFGLAK GP GD+THVST+VMGT GY AP+Y TG+LT KSD+YSFGV LL
Sbjct: 174 DAEFNAKLSDFGLAKAGPTGDRTHVSTQVMGTQGYAAPEYVATGRLTAKSDVYSFGVVLL 353
Query: 287 ELITGRKAIDHKKPAKEQNLVAWARPLFRDRRRFSEMIDPLLE 329
EL++GR+A+D +QNLV WARP D+RR ++D LE
Sbjct: 354 ELLSGRRAVDKTIAGVDQNLVDWARPYLGDKRRLFRIMDSKLE 482
>TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2, complete
Length = 2946
Score = 192 bits (487), Expect = 1e-49
Identities = 117/296 (39%), Positives = 171/296 (57%), Gaps = 1/296 (0%)
Frame = +1
Query: 72 DGKVAQTFTFAELAAATENFRADCFVGEGGFGKVYKGYLEKINQVVAIKQLDRNGVQGIR 131
D +A F F+ +++AT +F +GEGGFG VYKG L Q +A+K+L QG+
Sbjct: 1615 DIDLATIFDFSTISSATNHFSLSNKLGEGGFGPVYKGLLAN-GQEIAVKRLSNTSGQGME 1791
Query: 132 EFVVEVITLGLADHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLSPGQKPLDWNT 191
EF E+ + H NLVKL G ++ + M + L D S K +DWN
Sbjct: 1792 EFKNEIKLIARLQHRNLVKLFGCSVHQDENSHANKKMKI------LLD-STRSKLVDWNK 1950
Query: 192 RMKIAAGAARGLEYLHDKMKPPVIYRDLKCSNILLGEDYHSKLSDFGLAKVGPIGDKTHV 251
R++I G ARGL YLH + +I+RDLK SNILL ++ + K+SDFGLA++ IGD+
Sbjct: 1951 RLQIIDGIARGLLYLHQDSRLRIIHRDLKTSNILLDDEMNPKISDFGLARIF-IGDQVEA 2127
Query: 252 ST-RVMGTYGYCAPDYAMTGQLTFKSDIYSFGVALLELITGRKAIDHKKPAKEQNLVAWA 310
T RVMGTYGY P+YA+ G + KSD++SFGV +LE+I+G+K P NL++ A
Sbjct: 2128 RTKRVMGTYGYMPPEYAVHGSFSIKSDVFSFGVIVLEIISGKKIGRFYDPHHHLNLLSHA 2307
Query: 311 RPLFRDRRRFSEMIDPLLEGQYPVRGLYQALAIAAMCVQEQPNMRPVIADVVTALN 366
L+ + R E++D LL+ + + + +A +CVQ +P RP + +V LN
Sbjct: 2308 WRLWIEERPL-ELVDELLDDPVIPTEILRYIHVALLCVQRRPENRPDMLSIVLMLN 2472
>TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P14.15
{Arabidopsis thaliana;}, partial (37%)
Length = 1079
Score = 185 bits (469), Expect = 1e-47
Identities = 105/229 (45%), Positives = 146/229 (62%), Gaps = 7/229 (3%)
Frame = +3
Query: 136 EVITLGLADHPNLVKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLSPGQKPLDWNTRMKI 195
EV L DH NLVKLLGF +G +R+L+ EY+P G+L HL L K LD+N R++I
Sbjct: 6 EVELLAKIDHRNLVKLLGFIDKGNERILITEYVPNGTLREHLDGLRG--KILDFNQRLEI 179
Query: 196 AAGAARGLEYLHDKMKPPVIYRDLKCSNILLGEDYHSKLSDFGLAKVGPI-GDKTHVSTR 254
A A GL YLH + +I+RD+K SNILL E +K++DFG A++GP+ GD+TH+ST+
Sbjct: 180 AIDVAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFARLGPVNGDQTHISTK 359
Query: 255 VMGTYGYCAPDYAMTGQLTFKSDIYSFGVALLELITGRKAIDHKKPAKEQNLVAWARPLF 314
V GT GY P+Y T QLT KSD+YSFG+ LLE++TGR+ ++ KK A E+ + WA F
Sbjct: 360 VKGTVGYLDPEYMKTHQLTPKSDVYSFGILLLEILTGRRPVELKKAADERVTLRWA---F 530
Query: 315 RDRRRFS--EMIDPLLEGQYPVRGLYQALAIAAMCV----QEQPNMRPV 357
R S E++DPL+E L + L ++ C ++PNM+ V
Sbjct: 531 RKYNEGSVVELMDPLMEEAVNADVLMKMLDLSFQCAAPIRTDRPNMKSV 677
>TC12473 similar to UP|Q9M068 (Q9M068) Protein kinase-like protein, partial
(42%)
Length = 946
Score = 177 bits (449), Expect = 3e-45
Identities = 96/197 (48%), Positives = 128/197 (64%), Gaps = 9/197 (4%)
Frame = +3
Query: 60 VKNLNLNDGVSPDGKVAQTFTFAELAAATENFRADCFVGEGGFGKVYKGYLEKINQ---- 115
V N+ G + + F+F +L +AT++F+AD +GEGGFGKVYKG+L++
Sbjct: 363 VNNVKGGSGEFLEVAKLKVFSFGDLKSATKSFKADALIGEGGFGKVYKGWLDEKKLSPTK 542
Query: 116 -----VVAIKQLDRNGVQGIREFVVEVITLGLADHPNLVKLLGFCAEGEQRLLVYEYMPL 170
+VA+K+L+ +QG E+ E+ LG HPNLVKLLG+C + E+ LLVYE+MP
Sbjct: 543 PGSGIMVAVKKLNPESMQGFHEWQSEINFLGRISHPNLVKLLGYCRDDEEFLLVYEFMPR 722
Query: 171 GSLENHLHDLSPGQKPLDWNTRMKIAAGAARGLEYLHDKMKPPVIYRDLKCSNILLGEDY 230
GSLENHL + L WNTR+KIA GAARGL +LH K VIYRD K SNILL +Y
Sbjct: 723 GSLENHL--FRRNTESLSWNTRLKIAIGAARGLAFLHSLXK-IVIYRDFKASNILLDGNY 893
Query: 231 HSKLSDFGLAKVGPIGD 247
++K+S FGL K GP G+
Sbjct: 894 NAKISXFGLTKFGPSGE 944
>AV417846
Length = 429
Score = 177 bits (448), Expect = 4e-45
Identities = 88/144 (61%), Positives = 106/144 (73%)
Frame = +1
Query: 167 YMPLGSLENHLHDLSPGQKPLDWNTRMKIAAGAARGLEYLHDKMKPPVIYRDLKCSNILL 226
+MP GS+ENHL +P W+ R+KIA GAA+GL +LH +P VIYRD K SNILL
Sbjct: 1 FMPKGSMENHLFRRGSYFQPFSWSLRLKIALGAAKGLAFLHST-EPKVIYRDFKTSNILL 177
Query: 227 GEDYHSKLSDFGLAKVGPIGDKTHVSTRVMGTYGYCAPDYAMTGQLTFKSDIYSFGVALL 286
Y +KLSDFGLA+ GP+GDK+HVSTRVMGT GY AP+Y TG LT SD+YSFGV LL
Sbjct: 178 DTKYSAKLSDFGLARDGPVGDKSHVSTRVMGTRGYAAPEYLATGHLTANSDVYSFGVVLL 357
Query: 287 ELITGRKAIDHKKPAKEQNLVAWA 310
E+I+GR+AID + A E NLV WA
Sbjct: 358 EIISGRRAIDKNQLAGEHNLVEWA 429
>TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodulation aberrant
root formation protein), complete
Length = 3308
Score = 176 bits (446), Expect = 6e-45
Identities = 106/283 (37%), Positives = 154/283 (53%), Gaps = 6/283 (2%)
Frame = +2
Query: 89 ENFRADCFVGEGGFGKVYKGYLEKINQVVAIKQLDRNGVQGIREFVVEVITLGLADHPNL 148
E + + +G+GG G VY+G + V + + + + F E+ TLG H N+
Sbjct: 2186 ECLKEENIIGKGGAGIVYRGSMPNGTDVAIKRLVGQGSGRNDYGFRAEIETLGKIRHRNI 2365
Query: 149 VKLLGFCAEGEQRLLVYEYMPLGSLENHLHDLSPGQKPLDWNTRMKIAAGAARGLEYLHD 208
++LLG+ + + LL+YEYMP GSL LH G L W R KIA AARGL Y+H
Sbjct: 2366 MRLLGYVSNKDTNLLLYEYMPNGSLGEWLHGAKGGH--LRWEMRYKIAVEAARGLCYMHH 2539
Query: 209 KMKPPVIYRDLKCSNILLGEDYHSKLSDFGLAKVGPIGDKTHVSTRVMGTYGYCAPDYAM 268
P +I+RD+K +NILL D+ + ++DFGLAK + + + G+YGY AP+YA
Sbjct: 2540 DCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPEYAY 2719
Query: 269 TGQLTFKSDIYSFGVALLELITGRKAIDHKKPAKEQNLVAWARPLFRDRRRFSE------ 322
T ++ KSD+YSFGV LLELI GRK + + +V W + + S+
Sbjct: 2720 TLKVDEKSDVYSFGVVLLELIIGRKPVGEFGDGVD--IVGWVNKTMSELSQPSDTALVLA 2893
Query: 323 MIDPLLEGQYPVRGLYQALAIAAMCVQEQPNMRPVIADVVTAL 365
++DP L G YP+ + IA MCV+E RP + +VV L
Sbjct: 2894 VVDPRLSG-YPLTSVIHMFNIAMMCVKEMGPARPTMREVVHML 3019
>TC16403 homologue to UP|Q9LKY4 (Q9LKY4) Pti1 kinase-like protein, partial
(55%)
Length = 876
Score = 174 bits (440), Expect = 3e-44
Identities = 98/215 (45%), Positives = 132/215 (60%), Gaps = 5/215 (2%)
Frame = +3
Query: 187 LDWNTRMKIAAGAARGLEYLHDKMKPPVIYRDLKCSNILLGEDYHSKLSDFGLAKVGPIG 246
L W R+KIA GAARGLEYLH+K + +I+R +K SNILL +D +K++DF L+ P
Sbjct: 36 LSWTQRVKIAVGAARGLEYLHEKAETHIIHRYIKSSNILLFDDDVAKIADFDLSNQAPDA 215
Query: 247 DKTHVSTRVMGTYGYCAPDYAMTGQLTFKSDIYSFGVALLELITGRKAIDHKKPAKEQNL 306
STRV+GT+GY AP+YAMTGQLT KSD+YSFGV LLEL+TGRK +DH P +Q+L
Sbjct: 216 AARLHSTRVLGTFGYHAPEYAMTGQLTSKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSL 395
Query: 307 VAWARPLFRDRRRFSEMIDPLLEGQYPVRGLYQALAIAAMCVQEQPNMRPVIADVVTALN 366
V WA P + + + +D L+G+YPV+ + + A+AA+CVQ + TA +
Sbjct: 396 VTWATPKLSE-DKVKQCVDARLKGEYPVKSVAKMAAVAALCVQYEAEF--------TAKH 548
Query: 367 YLASQKYDPQIHHIQGSRKGS-----SSPRSRSER 396
S+ + S KGS SSP S R
Sbjct: 549 EHHSESSTASTEYSLNSSKGSAPHVNSSPSVPSSR 653
>TC10080 similar to UP|AAQ96340 (AAQ96340) Protein kinase-like protein,
partial (26%)
Length = 579
Score = 169 bits (427), Expect = 1e-42
Identities = 83/123 (67%), Positives = 97/123 (78%), Gaps = 1/123 (0%)
Frame = +1
Query: 287 ELITGRKAIDHKKPAKEQNLVAWARPLFRDRRRFSEMIDPLLEGQYPVRGLYQALAIAAM 346
ELIT RKAIDH KPAKEQNLVAW +PLFRD ++FS+M+DPLL+GQYP RGLYQALAIAAM
Sbjct: 1 ELITARKAIDHTKPAKEQNLVAWPKPLFRDSQKFSQMVDPLLQGQYPKRGLYQALAIAAM 180
Query: 347 CVQEQPNMRPVIADVVTALNYLASQKYDPQIHHIQGSRKGSSSPRSR-SERHRRVTSKDS 405
CVQEQP+MRPV+ADVVTALNYLAS KYDP H+Q S+ S R + HR +S +
Sbjct: 181 CVQEQPSMRPVVADVVTALNYLASNKYDPLNQHVQTSKNPSPPKDGRDDDDHRHSSSSEH 360
Query: 406 ETD 408
+
Sbjct: 361 NAE 369
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.321 0.139 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,533,432
Number of Sequences: 28460
Number of extensions: 82794
Number of successful extensions: 907
Number of sequences better than 10.0: 316
Number of HSP's better than 10.0 without gapping: 708
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 713
length of query: 412
length of database: 4,897,600
effective HSP length: 92
effective length of query: 320
effective length of database: 2,279,280
effective search space: 729369600
effective search space used: 729369600
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 56 (26.2 bits)
Medicago: description of AC135230.5


