

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC135229.3 - phase: 0
(781 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
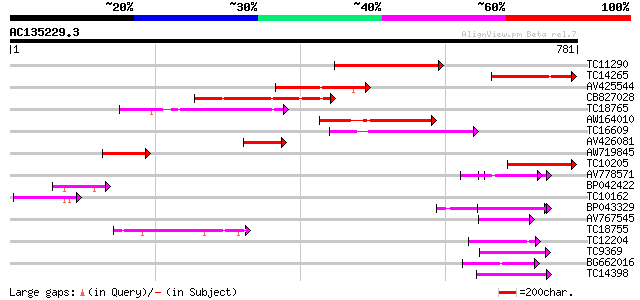
Score E
Sequences producing significant alignments: (bits) Value
TC11290 weakly similar to UP|AAN62760 (AAN62760) Disease resista... 196 2e-50
TC14265 weakly similar to UP|AAN62760 (AAN62760) Disease resista... 166 1e-41
AV425544 149 2e-36
CB827028 148 3e-36
TC18765 weakly similar to UP|Q9LVT1 (Q9LVT1) Disease resistance ... 131 5e-31
AW164010 123 1e-28
TC16609 weakly similar to PIR|T48468|T48468 disease resistance-l... 112 2e-25
AV426081 104 6e-23
AW719845 100 7e-22
TC10205 weakly similar to UP|Q8RL77 (Q8RL77) HpaG, partial (10%) 85 4e-17
AV778571 65 5e-11
BP042422 64 7e-11
TC10162 63 2e-10
BP043329 53 2e-07
AV767545 48 7e-06
TC18755 similar to UP|AAN85378 (AAN85378) Resistance protein (Fr... 47 9e-06
TC12204 weakly similar to UP|Q9ZSN3 (Q9ZSN3) NBS-LRR-like protei... 46 2e-05
TC9369 similar to UP|Q93X72 (Q93X72) Leucine-rich repeat resista... 46 3e-05
BG662016 45 3e-05
TC14398 similar to UP|Q96477 (Q96477) LRR protein, partial (86%) 44 8e-05
>TC11290 weakly similar to UP|AAN62760 (AAN62760) Disease resistance
protein-like protein MsR1, partial (18%)
Length = 459
Score = 196 bits (497), Expect = 2e-50
Identities = 101/152 (66%), Positives = 121/152 (79%), Gaps = 2/152 (1%)
Frame = +3
Query: 448 GIYQSTKEPFEQRKRLIIDMNNNKSG--LAEKQQGLMTRIFSKFMRLCVKQNPQQLAARI 505
GIYQS +EP +RKRL ID N +K L EKQQG+M+R SK RLC+KQ PQ + AR
Sbjct: 3 GIYQSIQEPVGKRKRLNIDTNEDKLEWLLQEKQQGIMSRTLSKIRRLCLKQKPQLVLART 182
Query: 506 LSVSTDETCALDWSHMQPAQVEVLILNIHTKQYSLPEWIAKMSKLRVLIITNYGFHPSKL 565
LS+STDETC D S +QPA+ EVLILN+ TK+YSLPE + +M+KL+ LI+TNYGFHPS+L
Sbjct: 183 LSISTDETCTPDLSLIQPAEAEVLILNLRTKKYSLPEILEEMNKLKALIVTNYGFHPSEL 362
Query: 566 NNIELLGSLQNLERIRLERISVPSFGTLKNLK 597
NN ELL SL NL+RIRLE+ISVPSFGTLKNLK
Sbjct: 363 NNFELLDSLSNLKRIRLEQISVPSFGTLKNLK 458
>TC14265 weakly similar to UP|AAN62760 (AAN62760) Disease resistance
protein-like protein MsR1, partial (5%)
Length = 766
Score = 166 bits (421), Expect = 1e-41
Identities = 82/117 (70%), Positives = 98/117 (83%)
Frame = +1
Query: 664 QDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNL 723
Q+IGKLENLELL LSSCTDL+ +P SIG+L L+ LDISNCISL SLPEE GNLCNLK+L
Sbjct: 1 QEIGKLENLELLRLSSCTDLKGLPDSIGRLSKLRLLDISNCISLPSLPEEIGNLCNLKSL 180
Query: 724 DMATCASIELPFSVVNLQNLKTITCDEETAATWEDFQHMLHNMKIEVLHVDVNLNWL 780
M +CA ELP S+VNLQNL T+ CDEETAA+WE F++++ ++KIEV VDVNLNWL
Sbjct: 181 YMTSCAGCELPSSIVNLQNL-TVVCDEETAASWEAFEYVIPHLKIEVPQVDVNLNWL 348
Score = 31.6 bits (70), Expect = 0.51
Identities = 25/76 (32%), Positives = 42/76 (54%)
Frame = +1
Query: 526 VEVLILNIHTKQYSLPEWIAKMSKLRVLIITNYGFHPSKLNNIELLGSLQNLERIRLERI 585
+E+L L+ T LP+ I ++SKLR+L I+N PS I L +L++L
Sbjct: 25 LELLRLSSCTDLKGLPDSIGRLSKLRLLDISNCISLPSLPEEIGNLCNLKSLYMTSCAGC 204
Query: 586 SVPSFGTLKNLKKLSL 601
+PS ++ NL+ L++
Sbjct: 205 ELPS--SIVNLQNLTV 246
Score = 31.2 bits (69), Expect = 0.66
Identities = 16/34 (47%), Positives = 19/34 (55%)
Frame = -3
Query: 683 LEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGN 716
L +P S GKL L D+S +S SLP E GN
Sbjct: 167 LHRLPISSGKLGRLMQXDMSRSLSFESLPIESGN 66
>AV425544
Length = 425
Score = 149 bits (375), Expect = 2e-36
Identities = 78/133 (58%), Positives = 94/133 (70%), Gaps = 3/133 (2%)
Frame = +1
Query: 367 ECFMDLALFPEDHRIPVAALVDMWAELYKLDDNGIQAMEIINKLGIMNLANVIIPRKNAS 426
ECFMDL LFPED RIPV AL+DMWAELY LD++G AM I+ L NL N I+ RK AS
Sbjct: 19 ECFMDLGLFPEDQRIPVTALIDMWAELYNLDEDGRNAMTIVLDLTSRNLINFIVTRKVAS 198
Query: 427 DTDNNNYNNHFIILHDILRELGIYQSTKEPFEQRKRLIIDMNNNKS---GLAEKQQGLMT 483
D YNNHF++LHD+LREL I+QS EPFEQRKRLIID+N + + + QQG +
Sbjct: 199 DA-GVCYNNHFVMLHDLLRELAIHQSKGEPFEQRKRLIIDLNGDNRPEWWVGQNQQGFIG 375
Query: 484 RIFSKFMRLCVKQ 496
R+FS R+ VKQ
Sbjct: 376 RLFSFLPRMLVKQ 414
>CB827028
Length = 572
Score = 148 bits (374), Expect = 3e-36
Identities = 90/197 (45%), Positives = 125/197 (62%), Gaps = 2/197 (1%)
Frame = +2
Query: 255 SRVAFPRFSTTC--ILKPLAHEDAVTLFHHYAQMEKNSSDIIDKNLVEKVVRSCNGLPLT 312
SR A R+ + +L+ L+ + A +LF H A + NS++I +LV+K+V+ C P+
Sbjct: 2 SRSAIARYDSGSPYVLERLSLDYATSLFLHSASLSPNSTEI-PVSLVKKIVKGCKRSPIA 178
Query: 313 IKVIATSLKNRPHDLWHKIVKELSQGHSILDSNTELLTRLQKIFDVLEDNPKIMECFMDL 372
+ V TSLKN+ +W K LS+G SIL+SN ++LT LQK DVL+ P +ECF DL
Sbjct: 179 LTVTGTSLKNKNLAVWRSRAKALSKGQSILESNHDVLTCLQKSLDVLD--PIAIECFKDL 352
Query: 373 ALFPEDHRIPVAALVDMWAELYKLDDNGIQAMEIINKLGIMNLANVIIPRKNASDTDNNN 432
LFPED RIP AALVDMWAEL DD AME I +L +N+A++ + RK A+D +
Sbjct: 353 GLFPEDQRIPAAALVDMWAELRCGDDE--TAMEKIYELVNLNMADIFVTRKVANDI--VD 520
Query: 433 YNNHFIILHDILRELGI 449
YN H++ H +LREL I
Sbjct: 521 YNYHYVAQHGLLRELAI 571
>TC18765 weakly similar to UP|Q9LVT1 (Q9LVT1) Disease resistance
protein-like, partial (18%)
Length = 688
Score = 131 bits (329), Expect = 5e-31
Identities = 86/242 (35%), Positives = 138/242 (56%), Gaps = 10/242 (4%)
Frame = +2
Query: 152 KTTLATKLCWDEEVNGKFKENIIFVTFSKTPMLKTIVERIHQH--------CGYSVPEFQ 203
KTTLA ++C D++V FKE I+F+T S++P ++ + +I H Y+VP++
Sbjct: 2 KTTLAREVCRDDQVRCHFKERILFLTVSQSPNVEELRAKIFGHIMGNRGLNANYAVPQWM 181
Query: 204 SDEDAVNKLGLLLKKVEGSPLLLVLDDVWPSSESLVEKLQFQMTGFKILVTSRVAFPR-F 262
+ ++ S +L+VLDDVW S ++E+L ++ G K LV SR F R F
Sbjct: 182 PQFECQSQ----------SQILVVLDDVW--SLPVLEQLVLRVPGCKYLVVSRFKFQRIF 325
Query: 263 STTCILKPLAHEDAVTLFHHYAQMEKNSSDIIDKNLVEKVVRSCNGLPLTIKVIATSLKN 322
+ T ++ L+ DA++LF H+A K+ ++NL+++VV C LPL +KVI SL++
Sbjct: 326 NDTYDVELLSEGDALSLFCHHAFGHKSIPFGANQNLIKQVVAECGRLPLALKVIGASLRD 505
Query: 323 RPHDLWHKIVKELSQGHSILDS-NTELLTRLQKIFDVLEDNPKIMECFMDLALFPEDHRI 381
+ W + LSQG SI +S L+ R+ + L + K+ ECF+DL FPED +I
Sbjct: 506 QNEMFWLSVKTRLSQGLSIGESYEVNLIDRMAISTNYLPE--KVKECFLDLCAFPEDKKI 679
Query: 382 PV 383
P+
Sbjct: 680 PL 685
>AW164010
Length = 494
Score = 123 bits (308), Expect = 1e-28
Identities = 64/161 (39%), Positives = 103/161 (63%)
Frame = +2
Query: 428 TDNNNYNNHFIILHDILRELGIYQSTKEPFEQRKRLIIDMNNNKSGLAEKQQGLMTRIFS 487
+D +YN H++ H +LR+L I Q+++E ++R RLII++N N
Sbjct: 26 SDIVDYNYHYVTQHGLLRDLAILQTSEETEDKRDRLIIEINRNN--------------LP 163
Query: 488 KFMRLCVKQNPQQLAARILSVSTDETCALDWSHMQPAQVEVLILNIHTKQYSLPEWIAKM 547
+ L +N +AARILS+STDE + + ++QP +VE L+LN+ K+Y+LP ++ KM
Sbjct: 164 AWWSL---ENEYPIAARILSISTDEAFSSELCNLQPTKVEALVLNLRGKKYTLPMFMEKM 334
Query: 548 SKLRVLIITNYGFHPSKLNNIELLGSLQNLERIRLERISVP 588
+KL+VL ITNY +P++L N E+L L NL+R+RLE++ +P
Sbjct: 335 NKLKVLTITNYDLYPAELENFEMLDHLSNLKRLRLEKVRIP 457
>TC16609 weakly similar to PIR|T48468|T48468 disease resistance-like protein
- Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(12%)
Length = 621
Score = 112 bits (280), Expect = 2e-25
Identities = 66/207 (31%), Positives = 104/207 (49%), Gaps = 2/207 (0%)
Frame = +1
Query: 441 HDILRELGIYQSTKEPFEQRKRLIIDMNNNKSGLAEKQQGLMTRIFSKFMRLCVKQNPQQ 500
HDILR+L + S + +R RL++ L ++ ++ Q
Sbjct: 43 HDILRDLALNLSNRGSINERLRLVMPKREGNGQLPKEW---------------LRYRGQP 177
Query: 501 LAARILSVSTDETCALDWSHMQPAQVEVLILNIHTKQYSLPEWIAKMSKLRVLIITNYGF 560
L ARI+S+ T E DW ++ + EVLILN + +Y LP +IA+M LR LI+ NY
Sbjct: 178 LEARIVSIHTGEMTEGDWCELEFPKAEVLILNFTSSEYFLPPFIARMPSLRALIVINYST 357
Query: 561 HPSKLNNIELLGSLQNLERIRLERISVPSFG--TLKNLKKLSLYMCNTILAFEKGSILIS 618
+ L+N+ + +L NL + LE++S P L+ L KL + +C + + ++
Sbjct: 358 TYAHLHNVSVFKNLSNLRSLWLEKVSTPQLSGIVLEKLGKLFIVLCQVNNSLKGKDANLA 537
Query: 619 DAFPNLEELNIDYCKDLVVLQTGICDI 645
FPNL E +D+C DL L + IC I
Sbjct: 538 HIFPNLSEFTLDHCDDLTELPSSICGI 618
>AV426081
Length = 179
Score = 104 bits (259), Expect = 6e-23
Identities = 49/59 (83%), Positives = 53/59 (89%)
Frame = +1
Query: 323 RPHDLWHKIVKELSQGHSILDSNTELLTRLQKIFDVLEDNPKIMECFMDLALFPEDHRI 381
RP++LW KIVKELSQGHS+LDS+TELLTRLQKI DV EDNP I ECFMDLALFPED RI
Sbjct: 1 RPYELWQKIVKELSQGHSVLDSHTELLTRLQKILDVAEDNPVIKECFMDLALFPEDQRI 177
>AW719845
Length = 209
Score = 100 bits (250), Expect = 7e-22
Identities = 45/67 (67%), Positives = 57/67 (84%)
Frame = +3
Query: 128 SKLKMELLRGGSSTLVLTGLGGLGKTTLATKLCWDEEVNGKFKENIIFVTFSKTPMLKTI 187
SKLKMELL+ G S + +TGLGGLGKTTLA +CWD+++ GKF+ENI+FVTFSKTP +K I
Sbjct: 6 SKLKMELLKDGVSVVAVTGLGGLGKTTLAAMVCWDKQIKGKFRENILFVTFSKTPNVKNI 185
Query: 188 VERIHQH 194
VER+ +H
Sbjct: 186 VERLFEH 206
>TC10205 weakly similar to UP|Q8RL77 (Q8RL77) HpaG, partial (10%)
Length = 586
Score = 85.1 bits (209), Expect = 4e-17
Identities = 42/96 (43%), Positives = 65/96 (66%), Gaps = 1/96 (1%)
Frame = +3
Query: 686 IPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMATCASI-ELPFSVVNLQNLK 744
+P SI L ++ LDIS+CISLS LPE+ G L L+ L++ C+ + ELP S+++L+NL+
Sbjct: 9 LPKSITNLEQVQFLDISDCISLSHLPEDMGELKKLEKLNVRGCSRLTELPTSIMDLENLE 188
Query: 745 TITCDEETAATWEDFQHMLHNMKIEVLHVDVNLNWL 780
+ CDEE A WE Q +L ++++ V D NL++L
Sbjct: 189 EVECDEEIAELWEPVQQILSDLQLHVAPTDFNLDFL 296
Score = 37.7 bits (86), Expect = 0.007
Identities = 23/54 (42%), Positives = 31/54 (56%), Gaps = 3/54 (5%)
Frame = +3
Query: 623 NLEE---LNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLE 673
NLE+ L+I C L L + ++ L+KLNV C +L+ LP I LENLE
Sbjct: 27 NLEQVQFLDISDCISLSHLPEDMGELKKLEKLNVRGCSRLTELPTSIMDLENLE 188
Score = 27.3 bits (59), Expect = 9.6
Identities = 29/109 (26%), Positives = 44/109 (39%), Gaps = 12/109 (11%)
Frame = -1
Query: 620 AFPNLEELNIDY------------CKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIG 667
A PN +E N++ C+ L + TG + S + + K S D+G
Sbjct: 328 ANPNYDERNLERKSKLKSVGATCSCRSLRICWTGSHN--SAISSSHSTSSKFSRSMIDVG 155
Query: 668 KLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGN 716
NLE S ++ P S GK L L ++S + S +FGN
Sbjct: 154 NSVNLEQPLTLSFSNFFNSPMSSGK*LRLMQSEMSRNCTCSRFVIDFGN 8
>AV778571
Length = 452
Score = 64.7 bits (156), Expect = 5e-11
Identities = 35/113 (30%), Positives = 63/113 (54%)
Frame = +3
Query: 622 PNLEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCT 681
PNLE+L+ Y ++L+ + + + L+ L++ C K+ + P KL +LE L LS C+
Sbjct: 84 PNLEKLSFVYSENLIKIHESVGFLNKLRILDIEGCCKIRTFPPM--KLASLEELYLSHCS 257
Query: 682 DLEAIPTSIGKLLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMATCASIELP 734
LE+ P + K+ N+ + + + + LP GNL L+ L++ C ++LP
Sbjct: 258 SLESFPEILAKMENITLIGVYHT-PIKELPSSIGNLTRLRRLELHGCGMLQLP 413
Score = 49.3 bits (116), Expect = 2e-06
Identities = 29/100 (29%), Positives = 49/100 (49%)
Frame = +3
Query: 647 SLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCIS 706
+L+KL+ L + + +G L L +L + C + P KL +L+ L +S+C S
Sbjct: 87 NLEKLSFVYSENLIKIHESVGFLNKLRILDIEGCCKIRTFPPM--KLASLEELYLSHCSS 260
Query: 707 LSSLPEEFGNLCNLKNLDMATCASIELPFSVVNLQNLKTI 746
L S PE + N+ + + ELP S+ NL L+ +
Sbjct: 261 LESFPEILAKMENITLIGVYHTPIKELPSSIGNLTRLRRL 380
Score = 44.3 bits (103), Expect = 8e-05
Identities = 29/94 (30%), Positives = 49/94 (51%), Gaps = 1/94 (1%)
Frame = +3
Query: 654 TNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISLSSLPEE 713
+ C ++ +P D+ L NLE LS +L I S+G L L+ LDI C + + P
Sbjct: 39 SECESITQIP-DLSGLPNLEKLSFVYSENLIKIHESVGFLNKLRILDIEGCCKIRTFPPM 215
Query: 714 FGNLCNLKNLDMATCASIE-LPFSVVNLQNLKTI 746
L +L+ L ++ C+S+E P + ++N+ I
Sbjct: 216 --KLASLEELYLSHCSSLESFPEILAKMENITLI 311
>BP042422
Length = 526
Score = 64.3 bits (155), Expect = 7e-11
Identities = 42/123 (34%), Positives = 59/123 (47%), Gaps = 43/123 (34%)
Frame = +3
Query: 59 HLDQPREEINSLIEES------------------------ACKSSFENDSYVVDENQSLI 94
HLD PREEI +LI E C ++ ++ V E Q+ +
Sbjct: 156 HLDPPREEIKTLIREKDAGEGDKCLCWGNFISLLQLCVNKLCHNNKDDFLNVDGEKQAQM 335
Query: 95 VNDVEETLYKAREILELLNHE-------------------NPKFTVGLDIPFSKLKMELL 135
NDV++TLYK R LEL++ E NP+FTVG+D+P +KLK+ELL
Sbjct: 336 ANDVKDTLYKVRGFLELISKEDFEQKFSGAPIKRPYGVPANPEFTVGVDVPLNKLKVELL 515
Query: 136 RGG 138
+ G
Sbjct: 516 KDG 524
>TC10162
Length = 577
Score = 63.2 bits (152), Expect = 2e-10
Identities = 42/118 (35%), Positives = 61/118 (51%), Gaps = 25/118 (21%)
Frame = +1
Query: 6 THIITLT--TRFNDVLNKISEIVETARKNQSSRLLFRLILKDLSPVVQDIKHYNEHLDQP 63
T ++ L RF ++L KIS++VE +RK+ ++ + R L + + VV +IK YNEHL QP
Sbjct: 211 TQLVALAPLARFQEILKKISQMVEKSRKSGIAQRILRSTLIEFNHVVLEIKRYNEHLAQP 390
Query: 64 REEINSLIEE----------SACKSSF-------------ENDSYVVDENQSLIVNDV 98
REEI +LI E C +S ++DS DE Q+L DV
Sbjct: 391 REEIKTLIREKDAGEEGEEGDKCVNSCLQVVTDRVGHGKDDSDSVDADEKQALTAKDV 564
>BP043329
Length = 484
Score = 52.8 bits (125), Expect = 2e-07
Identities = 35/104 (33%), Positives = 56/104 (53%), Gaps = 2/104 (1%)
Frame = +3
Query: 645 IISLKKLNVT-NCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISN 703
+ +LK LN++ N +P +IG L NLE++ L+ C + IP SIG L LK LD++
Sbjct: 27 LTTLKMLNLSYNPFYPGRIPPEIGNLTNLEVVWLTQCNLVGVIPDSIGNLKKLKDLDLAL 206
Query: 704 CISLSSLPEEFGNLCNLKNLDM-ATCASIELPFSVVNLQNLKTI 746
S+P L +L+ +++ S ELP + NL L+ +
Sbjct: 207 NDLYGSIPSSLTGLTSLRQIELYNNSLSGELPRGMGNLTELRLL 338
Score = 41.2 bits (95), Expect = 6e-04
Identities = 45/158 (28%), Positives = 72/158 (45%), Gaps = 2/158 (1%)
Frame = +3
Query: 588 PSFGTLKNLKKLSLYMCNTILAFEKGSILISDA-FPNLEELNIDYCKDLVVLQTGICDII 646
PS GTL LK L+L + F G I NLE + + C + V+ I ++
Sbjct: 12 PSLGTLTTLKMLNL----SYNPFYPGRIPPEIGNLTNLEVVWLTQCNLVGVIPDSIGNLK 179
Query: 647 SLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCIS 706
LK L++ S+P + L +L + L + + +P +G L L+ LD S
Sbjct: 180 KLKDLDLALNDLYGSIPSSLTGLTSLRQIELYNNSLSGELPRGMGNLTELRLLDASMNHL 359
Query: 707 LSSLPEEFGNLCNLKNLDM-ATCASIELPFSVVNLQNL 743
+PEE +L L++L++ ELP S+ + NL
Sbjct: 360 TGRIPEELCSL-PLESLNLYENRFEGELPASIADSPNL 470
>AV767545
Length = 562
Score = 47.8 bits (112), Expect = 7e-06
Identities = 29/76 (38%), Positives = 43/76 (56%)
Frame = -1
Query: 647 SLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCIS 706
SLK LN+ + + +P I L +LE L +S L AIP+S+G+LL L+ LD+S
Sbjct: 538 SLKVLNLGSNNISGPIPVSISNLIDLERLDISRNHILGAIPSSLGQLLELQWLDVSINSL 359
Query: 707 LSSLPEEFGNLCNLKN 722
S+P + NLK+
Sbjct: 358 AGSIPSSLSQITNLKH 311
>TC18755 similar to UP|AAN85378 (AAN85378) Resistance protein (Fragment),
complete
Length = 880
Score = 47.4 bits (111), Expect = 9e-06
Identities = 56/207 (27%), Positives = 98/207 (47%), Gaps = 19/207 (9%)
Frame = +3
Query: 144 LTGLGGLGKTTLATKLCWDEEVNGKFKENIIFVTFSKT----PMLKTIVERIHQHCGYSV 199
+TG+GG+GKTTL K +D+ V K ++T S++ +L+ + ++ V
Sbjct: 273 VTGMGGMGKTTL-VKQVYDDPVVIKHFRACAWITVSQSCEIGELLRDLARQLFSEIRRPV 449
Query: 200 PEFQSDEDAVNKLGLLLKK-VEGSPLLLVLDDVW--PSSESLVEKLQFQMTGFKILVTSR 256
P + ++L +++K ++ L+V DDVW E++ L G +I++T+R
Sbjct: 450 P-LGLENMRCDRLKMIIKDLLQRRRYLVVFDDVWHVREWEAVKYALPDNNCGSRIMITTR 626
Query: 257 VAFPRFSTTC-------ILKPLAHEDAVTLF-HHYAQMEKNSSDIIDKNLVEKVVRSCNG 308
+ F+++ L+PL ++A LF + S +I + ++R C G
Sbjct: 627 RSDLAFTSSTESKGKVYNLQPLKEDEAWELFCRKTFHGDSCPSHLI--GICTYILRKCEG 800
Query: 309 LPLTI----KVIATSLKNRPHDLWHKI 331
LPL I V+AT K R D W I
Sbjct: 801 LPLAIVAISGVLATKDKRR-IDEWDMI 878
>TC12204 weakly similar to UP|Q9ZSN3 (Q9ZSN3) NBS-LRR-like protein cD7
(Fragment), partial (3%)
Length = 328
Score = 46.2 bits (108), Expect = 2e-05
Identities = 32/100 (32%), Positives = 49/100 (49%), Gaps = 1/100 (1%)
Frame = +1
Query: 633 KDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGK 692
+ LVV + + SL L + NC S P+ + N+ L L C L++ P + K
Sbjct: 10 ESLVVTGVQLQYLQSLNSLRICNCPNFESFPEGGLRAPNMTNLHLEKCKKLKSFPQQMNK 189
Query: 693 -LLNLKHLDISNCISLSSLPEEFGNLCNLKNLDMATCASI 731
LL+L L+I C L S+PE G +L L++ CA +
Sbjct: 190 MLLSLMTLNIKECPELESIPEG-GFPDSLNLLEIFHCAKL 306
>TC9369 similar to UP|Q93X72 (Q93X72) Leucine-rich repeat resistance
protein-like protein, partial (70%)
Length = 859
Score = 45.8 bits (107), Expect = 3e-05
Identities = 32/98 (32%), Positives = 53/98 (53%), Gaps = 1/98 (1%)
Frame = +1
Query: 648 LKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDISNCISL 707
L +L++ N +P IG+L+ L++L+L +AIP IG+L +L HL +S
Sbjct: 13 LTRLDLHNNKLTGPIPPQIGRLKRLKILNLRWNKLQDAIPPEIGELKSLTHLYLSFNSFK 192
Query: 708 SSLPEEFGNLCNLKNLDMATCASI-ELPFSVVNLQNLK 744
+P+E NL +L+ L + I +P + LQNL+
Sbjct: 193 GEIPKELANLPDLRYLYLHENRLIGRIPPELGTLQNLR 306
>BG662016
Length = 459
Score = 45.4 bits (106), Expect = 3e-05
Identities = 39/108 (36%), Positives = 58/108 (53%), Gaps = 1/108 (0%)
Frame = +1
Query: 624 LEELNIDYCKDLVVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDL 683
LEELN ++ K + T ++I+LKKL V N +KL LP L L +L + L
Sbjct: 133 LEELNANFNKLSKLPDTIGFELINLKKLAV-NSNKLVLLPSSTSHLTALTVLD-ARLNCL 306
Query: 684 EAIPTSIGKLLNLKHLDIS-NCISLSSLPEEFGNLCNLKNLDMATCAS 730
A+P ++ L+NL+ L++S N L +LP G L + N +AT S
Sbjct: 307 RALPENLENLINLETLNVSQNFRYLDTLPYSIGLLLSWLNWMLATTIS 450
>TC14398 similar to UP|Q96477 (Q96477) LRR protein, partial (86%)
Length = 982
Score = 44.3 bits (103), Expect = 8e-05
Identities = 31/105 (29%), Positives = 52/105 (49%), Gaps = 1/105 (0%)
Frame = +1
Query: 643 CDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLNLKHLDIS 702
C S+ ++++ N + L D+G L +L+ L L IP +G L +L LD+
Sbjct: 313 CQDNSVTRVDLGNLNLSGHLVPDLGNLHSLQYLELYENNIQGTIPEELGNLQSLISLDLY 492
Query: 703 NCISLSSLPEEFGNLCNLKNLDM-ATCASIELPFSVVNLQNLKTI 746
+ S+P GNL NL+ L + + ++P S+ L NLK +
Sbjct: 493 HNNVSGSIPSSLGNLKNLRFLRLNNNHLTGQIPKSLSTLPNLKVL 627
Score = 40.0 bits (92), Expect = 0.001
Identities = 38/128 (29%), Positives = 55/128 (42%)
Frame = +1
Query: 576 NLERIRLERISVPSFGTLKNLKKLSLYMCNTILAFEKGSILISDAFPNLEELNIDYCKDL 635
+L + L VP G L +L+ L LY N I P EEL
Sbjct: 340 DLGNLNLSGHLVPDLGNLHSLQYLELYENN-----------IQGTIP--EELG------- 459
Query: 636 VVLQTGICDIISLKKLNVTNCHKLSSLPQDIGKLENLELLSLSSCTDLEAIPTSIGKLLN 695
++ SL L++ + + S+P +G L+NL L L++ IP S+ L N
Sbjct: 460 --------NLQSLISLDLYHNNVSGSIPSSLGNLKNLRFLRLNNNHLTGQIPKSLSTLPN 615
Query: 696 LKHLDISN 703
LK LD+SN
Sbjct: 616 LKVLDVSN 639
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.136 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,262,399
Number of Sequences: 28460
Number of extensions: 179976
Number of successful extensions: 970
Number of sequences better than 10.0: 94
Number of HSP's better than 10.0 without gapping: 901
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 940
length of query: 781
length of database: 4,897,600
effective HSP length: 97
effective length of query: 684
effective length of database: 2,136,980
effective search space: 1461694320
effective search space used: 1461694320
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC135229.3


