

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
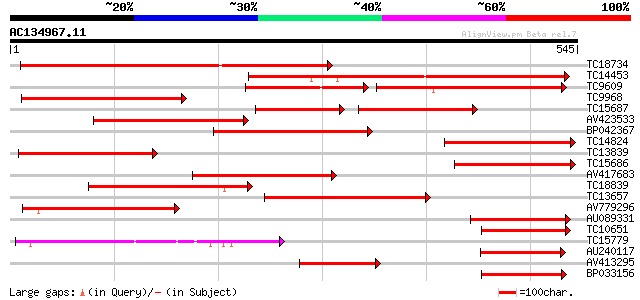
Query= AC134967.11 - phase: 0
(545 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC18734 similar to UP|Q8LFM3 (Q8LFM3) Pectinesterase-like protei... 404 e-113
TC14453 similar to PIR|T07634|T07634 pollen-specific protein hom... 294 2e-80
TC9609 similar to UP|Q39322 (Q39322) Bplo protein, partial (55%) 178 4e-80
TC9968 similar to UP|Q40473 (Q40473) PS60 protein precursor, par... 261 2e-70
TC15687 similar to UP|Q40473 (Q40473) PS60 protein precursor, pa... 160 3e-69
AV423533 245 1e-65
BP042367 218 2e-57
TC14824 similar to UP|Q93ZJ4 (Q93ZJ4) At1g76160/T23E18_10, parti... 209 9e-55
TC13839 homologue to UP|Q40473 (Q40473) PS60 protein precursor, ... 206 8e-54
TC15686 similar to UP|Q93ZJ4 (Q93ZJ4) At1g76160/T23E18_10, parti... 194 2e-50
AV417683 194 3e-50
TC18839 similar to GB|BAB01744.1|9280289|AP000603 l-ascorbate ox... 191 2e-49
TC13657 similar to UP|O23170 (O23170) Pectinesterase, partial (29%) 178 2e-45
AV779296 147 4e-36
AU089331 123 9e-29
TC10651 similar to UP|Q8LPS9 (Q8LPS9) AT5g66920/MUD21_18, partia... 120 7e-28
TC15779 similar to UP|O24093 (O24093) L-ascorbate oxidase precur... 114 3e-26
AU240117 114 5e-26
AV413295 110 8e-25
BP033156 107 4e-24
>TC18734 similar to UP|Q8LFM3 (Q8LFM3) Pectinesterase-like protein, partial
(47%)
Length = 929
Score = 404 bits (1039), Expect = e-113
Identities = 194/301 (64%), Positives = 234/301 (77%), Gaps = 1/301 (0%)
Frame = +3
Query: 11 LFLVTILLLFFTSVKCEDPYRFFTWKITYGDIYPLGVKQQGILINGQFPGPQIDAVTNEN 70
+F V + L+ + VK EDPY+F+TW +TYG + PLG QQ ILINGQFPGP++D VTN+N
Sbjct: 24 VFCVVLALVNVSLVKAEDPYKFYTWTVTYGTLSPLGSPQQVILINGQFPGPRLDLVTNDN 203
Query: 71 LIISVYNYLTEPFLISWNGIQHRRNSWQDGVSGTNCPIPPGKNFTYTLQVKDQIGTYFYF 130
+I+++ N L EPFL++WNGI+ R+NSWQDGV GTNCPIPP N+TY Q KDQIGTY YF
Sbjct: 204 VILNLINKLDEPFLLTWNGIKQRKNSWQDGVLGTNCPIPPNSNYTYKFQAKDQIGTYTYF 383
Query: 131 PSLGMHKAAGAFGGIRIWSRPRIPVPFPPPAGDFTLLAGDWSKLGHRRLRRVLENGHNLP 190
PS MHKA+G FG + + R IP+P+P P GDFTLL GDW K H+ L + L++G +L
Sbjct: 384 PSTKMHKASGGFGALNVLHRSVIPIPYPNPDGDFTLLIGDWYKTSHKTLSQTLDSGKSLA 563
Query: 191 FPDGLLINGRGWNGNTFTVDQGKTYRFRISNVGLATSINFRIQGHSLKLVEVEGSHTLQN 250
FPDGLLING+ +TFTV+QGKTY FRISNVGL+TSINFRIQGH+LKLVEVEGSHT+QN
Sbjct: 564 FPDGLLINGQA--HSTFTVNQGKTYMFRISNVGLSTSINFRIQGHTLKLVEVEGSHTIQN 737
Query: 251 TYSSLDIHLGQSYSVLVMANQPVKDYYIVVSTRFTRRVLTTTSILH-YSYSRIGVSGPVP 309
Y SLD+H+GQS SVLV NQP KDYYIV STRFT+ VLTTTS+LH S S S P P
Sbjct: 738 EYDSLDVHVGQSVSVLVTLNQPPKDYYIVASTRFTKTVLTTTSVLH*NSQSAASGSLPAP 917
Query: 310 P 310
P
Sbjct: 918 P 920
>TC14453 similar to PIR|T07634|T07634 pollen-specific protein homolog
T1P17.10 - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (56%)
Length = 1364
Score = 294 bits (753), Expect = 2e-80
Identities = 147/317 (46%), Positives = 206/317 (64%), Gaps = 8/317 (2%)
Frame = +2
Query: 230 FRIQGHSLKLVEVEGSHTLQNTYSSLDIHLGQSYSVLVMANQPVK-DYYIVVSTRFTRRV 288
FRIQ H+L L E EGS+T+Q Y+SLD+H+GQSY+ LV +Q DYYIV S RF
Sbjct: 2 FRIQNHNLLLAETEGSYTVQQNYTSLDVHVGQSYTFLVTMDQNASTDYYIVASARFVNES 181
Query: 289 ----LTTTSILHYSYSRIGVSGPVPPGPT--LDVVSSVFQARTIRWNLTASGPRPNPQGS 342
+T ILHY+ S+ GP+P P D S+ QAR+IRWN++ASG RPNPQGS
Sbjct: 182 RWQRVTGVGILHYTNSKGKARGPLPAAPEDQFDKTYSMNQARSIRWNVSASGARPNPQGS 361
Query: 343 YHYGLIKPTRTIMLANSAPY-INGKQRYAVNSVSYIAPDTPLKLADYFNIPGVFYVGGIS 401
+ YG I T +L N INGK+R ++ +S++ P TP++LAD F + ++ +
Sbjct: 362 FRYGSINVTEVYVLKNKPRVKINGKKRATLSGISFVNPSTPIRLADQFKLKDIYKLD-FP 538
Query: 402 TSPTGGNAYLQTAVMGANFHEYVEIVFQNWENSVQSWHIDGYSFFVVGFGSGQWTPNSRG 461
T P G+ +T+V+ ++ ++EI+ QN + + ++H+DGY+FFVVG G W+ NSRG
Sbjct: 539 TRPLTGSPRAETSVINGSYRGFIEIILQNNDTRMHTYHLDGYAFFVVGMDYGDWSDNSRG 718
Query: 462 RYNLRDTVARCTTQVYPRSWTAIYMALDNVGMWNIRSENWERQYLGQQFYLRVYTPSKSL 521
YN D +AR TTQVYP +WTAI ++LDNVG+WN+R+EN + YLGQ+ Y+RV P +
Sbjct: 719 TYNKWDGIARATTQVYPGAWTAILVSLDNVGIWNLRTENLDSWYLGQETYIRVVNPEPNN 898
Query: 522 RDEYPVPKNALLCGRAS 538
+ E P+P NAL CG S
Sbjct: 899 KTELPIPDNALFCGALS 949
>TC9609 similar to UP|Q39322 (Q39322) Bplo protein, partial (55%)
Length = 1104
Score = 178 bits (451), Expect(2) = 4e-80
Identities = 89/186 (47%), Positives = 119/186 (63%), Gaps = 3/186 (1%)
Frame = +2
Query: 353 TIMLANSAPYINGKQRYAVNSVSYIAPDTPLKLADYFNIPG-VFYVGGISTSPTG--GNA 409
TI L N+ GK RYA+N VS++ +TPLKLA+YF + VF IS P
Sbjct: 392 TIKLVNTVSKAQGKLRYAINGVSHVDGETPLKLAEYFGVADKVFKYNIISDEPPADLNTI 571
Query: 410 YLQTAVMGANFHEYVEIVFQNWENSVQSWHIDGYSFFVVGFGSGQWTPNSRGRYNLRDTV 469
L V+ A F ++ I+F+ +VQS+++DGYSFF+V G+WTP R YNL DTV
Sbjct: 572 TLAPNVVNATFRTFIXIIFEKPGKTVQSYNLDGYSFFLVAVEPGKWTPEKRKNYNLLDTV 751
Query: 470 ARCTTQVYPRSWTAIYMALDNVGMWNIRSENWERQYLGQQFYLRVYTPSKSLRDEYPVPK 529
+R T QV+P+SW AI + DN GMWN+RSE+ E +YLGQQ Y+ V +P +SLRDEY +
Sbjct: 752 SRHTVQVFPKSWAAIMLTFDNAGMWNLRSEHAENRYLGQQLYISVLSPERSLRDEYNILD 931
Query: 530 NALLCG 535
+ LLCG
Sbjct: 932 SQLLCG 949
Score = 137 bits (346), Expect(2) = 4e-80
Identities = 67/119 (56%), Positives = 86/119 (71%)
Frame = +3
Query: 227 SINFRIQGHSLKLVEVEGSHTLQNTYSSLDIHLGQSYSVLVMANQPVKDYYIVVSTRFTR 286
S+NFRIQGH +KLVE EGSH +QNTY SLD+H+GQ Y+VLV A++ KDYY+V STRFT+
Sbjct: 15 SLNFRIQGHPMKLVETEGSHVVQNTYDSLDVHVGQCYTVLVTADKDPKDYYLVASTRFTK 194
Query: 287 RVLTTTSILHYSYSRIGVSGPVPPGPTLDVVSSVFQARTIRWNLTASGPRPNPQGSYHY 345
LT I+ Y+ + +G + P P + S+ Q R+ RWNLTAS +PNPQGSYHY
Sbjct: 195 YSLTGKGIIRYT-NGVGPASPELPPAPVGWAWSLNQFRSFRWNLTASAAKPNPQGSYHY 368
>TC9968 similar to UP|Q40473 (Q40473) PS60 protein precursor, partial (27%)
Length = 631
Score = 261 bits (667), Expect = 2e-70
Identities = 120/159 (75%), Positives = 135/159 (84%)
Frame = +2
Query: 12 FLVTILLLFFTSVKCEDPYRFFTWKITYGDIYPLGVKQQGILINGQFPGPQIDAVTNENL 71
F T+LLL V EDPYRFF W ITYGDIYPLGVKQQGILINGQFPGP+I +VTN+NL
Sbjct: 155 FCSTLLLLNLVHVSAEDPYRFFDWTITYGDIYPLGVKQQGILINGQFPGPEIYSVTNDNL 334
Query: 72 IISVYNYLTEPFLISWNGIQHRRNSWQDGVSGTNCPIPPGKNFTYTLQVKDQIGTYFYFP 131
+I+V+N+L EPFL+SWNG+Q RRNS+QDGV GT CPIPPGKNFTY LQVKDQIG++FYFP
Sbjct: 335 VINVHNHLPEPFLLSWNGVQQRRNSYQDGVYGTTCPIPPGKNFTYNLQVKDQIGSFFYFP 514
Query: 132 SLGMHKAAGAFGGIRIWSRPRIPVPFPPPAGDFTLLAGD 170
SL HKAAG FG I+I SRPRIPVPFP PA DF+LL GD
Sbjct: 515 SLAFHKAAGGFGAIKILSRPRIPVPFPEPAADFSLLIGD 631
>TC15687 similar to UP|Q40473 (Q40473) PS60 protein precursor, partial (37%)
Length = 669
Score = 160 bits (404), Expect(2) = 3e-69
Identities = 78/114 (68%), Positives = 91/114 (79%)
Frame = +3
Query: 336 RPNPQGSYHYGLIKPTRTIMLANSAPYINGKQRYAVNSVSYIAPDTPLKLADYFNIPGVF 395
RPNPQGSYHYGLI +RTI L +SA +N KQRYA+NSVS+ DTPLKLADYFNI GVF
Sbjct: 258 RPNPQGSYHYGLINISRTIKLESSAAQVNNKQRYAINSVSFTPADTPLKLADYFNIGGVF 437
Query: 396 YVGGISTSPTGGNAYLQTAVMGANFHEYVEIVFQNWENSVQSWHIDGYSFFVVG 449
+G I SP+G YL T+VMGA+F +VEIVFQN E+ VQS+HIDGYSF+VVG
Sbjct: 438 QLGSIPDSPSGRPMYLDTSVMGADFRAFVEIVFQNHESIVQSYHIDGYSFWVVG 599
Score = 119 bits (298), Expect(2) = 3e-69
Identities = 58/86 (67%), Positives = 70/86 (80%)
Frame = +1
Query: 237 LKLVEVEGSHTLQNTYSSLDIHLGQSYSVLVMANQPVKDYYIVVSTRFTRRVLTTTSILH 296
+KLVEVEG+HT+Q TYSS+D+H+GQSYSVLV A+Q KDYYIV STRFT RVLT+T ILH
Sbjct: 1 MKLVEVEGTHTVQTTYSSIDVHVGQSYSVLVTADQAPKDYYIVASTRFTNRVLTSTGILH 180
Query: 297 YSYSRIGVSGPVPPGPTLDVVSSVFQ 322
YS S+ VSGP+P GPT + S+ Q
Sbjct: 181 YSNSQQSVSGPIPGGPTTQIDWSLEQ 258
>AV423533
Length = 472
Score = 245 bits (626), Expect = 1e-65
Identities = 112/149 (75%), Positives = 125/149 (83%)
Frame = -2
Query: 81 EPFLISWNGIQHRRNSWQDGVSGTNCPIPPGKNFTYTLQVKDQIGTYFYFPSLGMHKAAG 140
EPFL +WNG+ RRNSWQDGV GTNCPIPPG+NFTY LQVKDQIG+YFYFPSL H+AAG
Sbjct: 471 EPFLFTWNGVFQRRNSWQDGVFGTNCPIPPGQNFTYVLQVKDQIGSYFYFPSLAFHRAAG 292
Query: 141 AFGGIRIWSRPRIPVPFPPPAGDFTLLAGDWSKLGHRRLRRVLENGHNLPFPDGLLINGR 200
+GGI+I SRP IPVPFPPPAGDFT+LAGDW K H LR +L++G +LPFPDGLLING
Sbjct: 291 GYGGIKIASRPLIPVPFPPPAGDFTILAGDWYKRNHTDLRAILDSGSDLPFPDGLLINGH 112
Query: 201 GWNGNTFTVDQGKTYRFRISNVGLATSIN 229
G N TF V+QG TYRFRISNVGL TSIN
Sbjct: 111 GSNAYTFMVNQGNTYRFRISNVGLTTSIN 25
>BP042367
Length = 465
Score = 218 bits (555), Expect = 2e-57
Identities = 109/152 (71%), Positives = 124/152 (80%)
Frame = -3
Query: 197 INGRGWNGNTFTVDQGKTYRFRISNVGLATSINFRIQGHSLKLVEVEGSHTLQNTYSSLD 256
INGRG G + V+QGKTYR RISNVGL S+NFRIQ H +KLVEVEG+HTLQ TYSSLD
Sbjct: 463 INGRGSGGASLNVEQGKTYRLRISNVGLEHSLNFRIQNHKMKLVEVEGTHTLQTTYSSLD 284
Query: 257 IHLGQSYSVLVMANQPVKDYYIVVSTRFTRRVLTTTSILHYSYSRIGVSGPVPPGPTLDV 316
+H+GQSYSVLV A+QP +DYYIVVSTRFT VLT+T +L YS S VSGP P GPT+ V
Sbjct: 283 VHVGQSYSVLVTADQPAQDYYIVVSTRFTSPVLTSTGVLRYSNSAGQVSGPPPGGPTIQV 104
Query: 317 VSSVFQARTIRWNLTASGPRPNPQGSYHYGLI 348
S+ QAR+IR NLTASGPRPNPQGSYHYG+I
Sbjct: 103 DWSLNQARSIRTNLTASGPRPNPQGSYHYGMI 8
>TC14824 similar to UP|Q93ZJ4 (Q93ZJ4) At1g76160/T23E18_10, partial (23%)
Length = 682
Score = 209 bits (532), Expect = 9e-55
Identities = 97/126 (76%), Positives = 108/126 (84%)
Frame = +3
Query: 419 NFHEYVEIVFQNWENSVQSWHIDGYSFFVVGFGSGQWTPNSRGRYNLRDTVARCTTQVYP 478
+F +VEIVFQN EN VQS+H+DGYSFFVVG G WT +SR +YNLRD VARCTTQVYP
Sbjct: 3 DFRSFVEIVFQNNENIVQSYHLDGYSFFVVGMDGGTWTASSRNQYNLRDAVARCTTQVYP 182
Query: 479 RSWTAIYMALDNVGMWNIRSENWERQYLGQQFYLRVYTPSKSLRDEYPVPKNALLCGRAS 538
SWTAIY+ALDNVGMWN+R+E W RQYLGQQFYLRVYT S S+RDE+PVPKNALLCGRAS
Sbjct: 183 YSWTAIYVALDNVGMWNLRTEFWARQYLGQQFYLRVYTASTSIRDEFPVPKNALLCGRAS 362
Query: 539 GRVTRP 544
GR RP
Sbjct: 363 GRHKRP 380
>TC13839 homologue to UP|Q40473 (Q40473) PS60 protein precursor, partial
(22%)
Length = 494
Score = 206 bits (524), Expect = 8e-54
Identities = 91/134 (67%), Positives = 112/134 (82%)
Frame = +1
Query: 9 SSLFLVTILLLFFTSVKCEDPYRFFTWKITYGDIYPLGVKQQGILINGQFPGPQIDAVTN 68
++L L + F + EDPY+FF+W +TYGDIYPLGV+Q+GILINGQFPGP I +VTN
Sbjct: 91 ATLLLCFFAISLFQLTQAEDPYKFFSWNVTYGDIYPLGVRQRGILINGQFPGPDIHSVTN 270
Query: 69 ENLIISVYNYLTEPFLISWNGIQHRRNSWQDGVSGTNCPIPPGKNFTYTLQVKDQIGTYF 128
+NLII+V+N L EPFL+SWNGIQ+RRNS++DGV GT CPIPPG+NFTY LQVKDQIG+++
Sbjct: 271 DNLIINVFNSLDEPFLLSWNGIQNRRNSFEDGVFGTTCPIPPGRNFTYILQVKDQIGSFY 450
Query: 129 YFPSLGMHKAAGAF 142
YFPSL HKAAG F
Sbjct: 451 YFPSLAFHKAAGGF 492
>TC15686 similar to UP|Q93ZJ4 (Q93ZJ4) At1g76160/T23E18_10, partial (22%)
Length = 666
Score = 194 bits (494), Expect = 2e-50
Identities = 89/117 (76%), Positives = 100/117 (85%)
Frame = +3
Query: 428 FQNWENSVQSWHIDGYSFFVVGFGSGQWTPNSRGRYNLRDTVARCTTQVYPRSWTAIYMA 487
FQN E+ VQS+HIDGYSF+VVG G+WTP SR YNL D V+R TTQVYP+SWTAIYMA
Sbjct: 3 FQNHESIVQSYHIDGYSFWVVGMDGGEWTPASRNEYNLIDAVSRSTTQVYPKSWTAIYMA 182
Query: 488 LDNVGMWNIRSENWERQYLGQQFYLRVYTPSKSLRDEYPVPKNALLCGRASGRVTRP 544
LDNVGMWNIRS W RQYLGQQFYLRVY+P S+RDEYP+PKNALLCG+A+GR TRP
Sbjct: 183 LDNVGMWNIRSGFWARQYLGQQFYLRVYSPVGSIRDEYPIPKNALLCGKAAGRATRP 353
>AV417683
Length = 427
Score = 194 bits (493), Expect = 3e-50
Identities = 94/139 (67%), Positives = 111/139 (79%)
Frame = +3
Query: 176 HRRLRRVLENGHNLPFPDGLLINGRGWNGNTFTVDQGKTYRFRISNVGLATSINFRIQGH 235
H L+ L+ G+ LP PDG+LINGRG NG +F V+QGKTYR RISNVGL S+NFRIQ H
Sbjct: 6 HTTLKAHLDKGNKLPIPDGVLINGRGPNGVSFNVEQGKTYRLRISNVGLQHSLNFRIQNH 185
Query: 236 SLKLVEVEGSHTLQNTYSSLDIHLGQSYSVLVMANQPVKDYYIVVSTRFTRRVLTTTSIL 295
+KLVEVEG+HTLQ TYSSLD+H+GQSYSVLV A+QP +DYYIV STRF+ ++LTTT IL
Sbjct: 186 KMKLVEVEGTHTLQTTYSSLDVHVGQSYSVLVTADQPAQDYYIVASTRFSSKILTTTGIL 365
Query: 296 HYSYSRIGVSGPVPPGPTL 314
HYS S V GP P GPT+
Sbjct: 366 HYSNSAGPVKGPPPGGPTI 422
>TC18839 similar to GB|BAB01744.1|9280289|AP000603 l-ascorbate oxidase;
pectinesterase-like protein; pollen-specific
protein-like {Arabidopsis thaliana;} , partial (29%)
Length = 498
Score = 191 bits (486), Expect = 2e-49
Identities = 85/163 (52%), Positives = 119/163 (72%), Gaps = 5/163 (3%)
Frame = +1
Query: 76 YNYLTEPFLISWNGIQHRRNSWQDGVSGTNCPIPPGKNFTYTLQVKDQIGTYFYFPSLGM 135
+N L EP +W+G+Q R+NSWQDGV+GT+CPI PG N+TY QVKDQIG+YFY+P+ +
Sbjct: 10 FNNLDEPLQFTWHGVQQRKNSWQDGVAGTSCPILPGTNYTYHFQVKDQIGSYFYYPTTAL 189
Query: 136 HKAAGAFGGIRIWSRPRIPVPFPPPAGDFTLLAGDWSKLGHRRLRRVLENGHNLPFPDGL 195
H+AAG FGG+RI+SR IPVP+P P ++ ++ GDW H+ L+ L++G ++ PDG+
Sbjct: 190 HRAAGGFGGLRIFSRLLIPVPYPDPEDEYWVIIGDWYAKSHKTLKSFLDSGRSIGRPDGV 369
Query: 196 LINGRGWNGN-----TFTVDQGKTYRFRISNVGLATSINFRIQ 233
LING+ G+ +T+ GKTY++RI NVGL S+NFRIQ
Sbjct: 370 LINGKTAKGDGSDQPLYTMKPGKTYKYRICNVGLKDSLNFRIQ 498
>TC13657 similar to UP|O23170 (O23170) Pectinesterase, partial (29%)
Length = 498
Score = 178 bits (452), Expect = 2e-45
Identities = 91/159 (57%), Positives = 111/159 (69%)
Frame = +3
Query: 246 HTLQNTYSSLDIHLGQSYSVLVMANQPVKDYYIVVSTRFTRRVLTTTSILHYSYSRIGVS 305
HTLQ +Y SLD+H+GQS +VLV + V DYYIV STRFT +L+ T L Y S S
Sbjct: 3 HTLQQSYESLDVHVGQSVTVLVTLSGSVSDYYIVASTRFTDPILSITGTLRYVGSNSKAS 182
Query: 306 GPVPPGPTLDVVSSVFQARTIRWNLTASGPRPNPQGSYHYGLIKPTRTIMLANSAPYING 365
GP+P GP + S+ QAR+IR NLTA+ RPNPQGS+HYG I TRT++LANS +NG
Sbjct: 183 GPLPIGPADGIEWSMKQARSIRLNLTANAARPNPQGSFHYGTIPITRTLVLANSKTLLNG 362
Query: 366 KQRYAVNSVSYIAPDTPLKLADYFNIPGVFYVGGISTSP 404
K RYAVN +SY+ P TPLKLAD+FNI GVF I+ P
Sbjct: 363 KVRYAVNGISYVNPSTPLKLADWFNISGVFDFSTITDVP 479
>AV779296
Length = 549
Score = 147 bits (371), Expect = 4e-36
Identities = 73/153 (47%), Positives = 100/153 (64%), Gaps = 2/153 (1%)
Frame = +3
Query: 13 LVTILLLFFTSVKC--EDPYRFFTWKITYGDIYPLGVKQQGILINGQFPGPQIDAVTNEN 70
L+++L + S C DP +++Y + PLG+ Q+ I ING FPGP I+ TN +
Sbjct: 90 LLSLLHMALLSSLCLAGDPTVLADLRVSYTTVSPLGIPQRVIAINGDFPGPVINVTTNNH 269
Query: 71 LIISVYNYLTEPFLISWNGIQHRRNSWQDGVSGTNCPIPPGKNFTYTLQVKDQIGTYFYF 130
+ ++V+N L E LI+W IQ RR++WQDGV GTNCPIPP N+TY QVKDQ G++FYF
Sbjct: 270 VSVNVHNELDEELLITWPEIQMRRDAWQDGVLGTNCPIPPKWNWTYKFQVKDQXGSFFYF 449
Query: 131 PSLGMHKAAGAFGGIRIWSRPRIPVPFPPPAGD 163
PSL +A+G FG + I +R IP+PF GD
Sbjct: 450 PSLKFQRASGGFGPLVINNRQIIPIPFAQADGD 548
>AU089331
Length = 602
Score = 123 bits (308), Expect = 9e-29
Identities = 55/96 (57%), Positives = 68/96 (70%)
Frame = +1
Query: 444 SFFVVGFGSGQWTPNSRGRYNLRDTVARCTTQVYPRSWTAIYMALDNVGMWNIRSENWER 503
SFF V G WTP R YNL D V+R T QV+P+SW AI + DNVG+WN+RSE E
Sbjct: 10 SFFYVAVEPGTWTPEKRKTYNLLDAVSRHTVQVFPKSWAAIMLTFDNVGVWNLRSEIAEN 189
Query: 504 QYLGQQFYLRVYTPSKSLRDEYPVPKNALLCGRASG 539
+YLGQQ Y+ V TP +SLRDEY +P+NAL+CG+ G
Sbjct: 190 RYLGQQLYISVMTPERSLRDEYNIPENALICGQIKG 297
>TC10651 similar to UP|Q8LPS9 (Q8LPS9) AT5g66920/MUD21_18, partial (16%)
Length = 479
Score = 120 bits (300), Expect = 7e-28
Identities = 55/86 (63%), Positives = 64/86 (73%)
Frame = +3
Query: 454 QWTPNSRGRYNLRDTVARCTTQVYPRSWTAIYMALDNVGMWNIRSENWERQYLGQQFYLR 513
QWT SR YNL D + R T QVYP SWTAI ++LDN GMWN+RS WERQYLGQQ YLR
Sbjct: 3 QWTDASRSTYNLVDALTRHTAQVYPNSWTAILVSLDNQGMWNLRSAIWERQYLGQQVYLR 182
Query: 514 VYTPSKSLRDEYPVPKNALLCGRASG 539
V+ +SL +EY +P NALLCG+A G
Sbjct: 183 VWNAQQSLANEYGIPNNALLCGKAIG 260
>TC15779 similar to UP|O24093 (O24093) L-ascorbate oxidase precursor,
partial (50%)
Length = 1304
Score = 114 bits (286), Expect = 3e-26
Identities = 97/293 (33%), Positives = 137/293 (46%), Gaps = 34/293 (11%)
Frame = +1
Query: 6 NVVSSLFLVTILL--LFFTSVKCEDPYRFFTWKITYGDIYPLGVKQQGILINGQFPGPQI 63
+++ +LFL +L LF +SV R + + + Y P G++ + INGQFPGP I
Sbjct: 115 SLLKALFLWCLLWVGLFQSSVVVGAKVRHYKFDVEYMFKKPDGLEHVVMGINGQFPGPTI 294
Query: 64 DAVTNENLIISVYNYL-TEPFLISWNGIQHRRNSWQDGVSG-TNCPIPPGKNFTYTLQVK 121
A + L I++ N L TE +I +GI+ W DG + + C I PG+ F Y V
Sbjct: 295 KAEVGDTLDIALTNKLHTEGTVIHGHGIRQLGTPWADGTAAISQCAINPGETFHYWFTV- 471
Query: 122 DQIGTYFYFPSLGMHKAAGAFGGIRIWSRPRIPVPFPPPAGDFTLLAGD-WSKLGHRRLR 180
D+ GTYFY GM +AAG +G + + PF G+F LL D W K H +
Sbjct: 472 DRPGTYFYHGHYGMQRAAGLYGSLIVDLPKGQNEPFHYD-GEFNLLLSDLWHKSSHE--Q 642
Query: 181 RVLENGHNLPF---PDGLLINGRGWNG--------NTFTVDQ------------------ 211
V + L + P LLINGRG NT V Q
Sbjct: 643 EVGLSSKPLKWIGEPQSLLINGRGQFNCSLAAKFVNTSLVPQCQFKGGEECAPEILHVEP 822
Query: 212 GKTYRFRISNVGLATSINFRIQGHSLKLVEVEGSHTLQNTYSSLDIHLGQSYS 264
KTYR RI++ S+N I H L +VE +G+H +DI+ G++YS
Sbjct: 823 NKTYRIRIASTTSMASLNLAISNHKLIVVEADGNHVQPFAVDDIDIYSGETYS 981
>AU240117
Length = 300
Score = 114 bits (284), Expect = 5e-26
Identities = 48/82 (58%), Positives = 61/82 (73%)
Frame = +2
Query: 453 GQWTPNSRGRYNLRDTVARCTTQVYPRSWTAIYMALDNVGMWNIRSENWERQYLGQQFYL 512
G+WTP R YNL DTV+R T Q+YP+SW AI + DN GMWN+RSE+ E +YLGQQ Y+
Sbjct: 53 GKWTPEKRKNYNLLDTVSRHTVQIYPKSWAAIMLTFDNAGMWNLRSEHTENRYLGQQLYI 232
Query: 513 RVYTPSKSLRDEYPVPKNALLC 534
V +P +SLRDEY +P+ LLC
Sbjct: 233 SVLSPERSLRDEYNIPETQLLC 298
>AV413295
Length = 236
Score = 110 bits (274), Expect = 8e-25
Identities = 55/78 (70%), Positives = 62/78 (78%)
Frame = +3
Query: 279 VVSTRFTRRVLTTTSILHYSYSRIGVSGPVPPGPTLDVVSSVFQARTIRWNLTASGPRPN 338
V STRFT RVLT+T ILHYS S+ VSGP+P GPT + S+ QAR+IR NLTASGPRPN
Sbjct: 3 VASTRFTNRVLTSTGILHYSNSQQSVSGPIPGGPTTQIDWSLEQARSIRTNLTASGPRPN 182
Query: 339 PQGSYHYGLIKPTRTIML 356
PQGSYHYGLI +RTI L
Sbjct: 183 PQGSYHYGLINISRTIKL 236
>BP033156
Length = 408
Score = 107 bits (268), Expect = 4e-24
Identities = 47/82 (57%), Positives = 60/82 (72%)
Frame = -3
Query: 454 QWTPNSRGRYNLRDTVARCTTQVYPRSWTAIYMALDNVGMWNIRSENWERQYLGQQFYLR 513
+WTP R YNL DTV+R T QV+P+SW AI + DN GMWN+RSE+ E +YLGQQ Y+
Sbjct: 406 KWTPEKRKNYNLLDTVSRHTVQVFPQSWAAIMLTFDNAGMWNLRSEHAENRYLGQQLYIS 227
Query: 514 VYTPSKSLRDEYPVPKNALLCG 535
V +P +SLRDEY + + LLCG
Sbjct: 226 VLSPERSLRDEYNILDSQLLCG 161
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.321 0.139 0.441
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,749,350
Number of Sequences: 28460
Number of extensions: 168755
Number of successful extensions: 897
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 873
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 877
length of query: 545
length of database: 4,897,600
effective HSP length: 95
effective length of query: 450
effective length of database: 2,193,900
effective search space: 987255000
effective search space used: 987255000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 57 (26.6 bits)
Medicago: description of AC134967.11


