

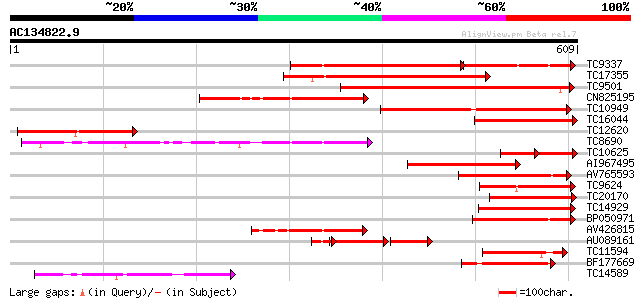
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC134822.9 + phase: 0
(609 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC9337 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B pre... 232 5e-98
TC17355 similar to UP|Q9FF77 (Q9FF77) Pectinesterase, partial (34%) 262 1e-70
TC9501 weakly similar to UP|Q9MBB6 (Q9MBB6) Pectin methylesteras... 256 1e-68
CN825195 253 8e-68
TC10949 similar to UP|PME3_PHAVU (Q43111) Pectinesterase 3 precu... 221 3e-58
TC16044 similar to UP|CAE76634 (CAE76634) Pectin methylesterase ... 194 5e-50
TC12620 weakly similar to UP|Q9FK05 (Q9FK05) Pectinesterase, par... 182 2e-46
TC8690 similar to UP|Q8RVX1 (Q8RVX1) Pectin methylesterase precu... 172 2e-43
TC10625 similar to UP|CAE76634 (CAE76634) Pectin methylesterase ... 92 2e-36
AI967495 141 3e-34
AV765593 141 3e-34
TC9624 similar to UP|Q43234 (Q43234) Pectinmethylesterase precur... 127 5e-30
TC20170 similar to UP|Q9M5J0 (Q9M5J0) Pectin methylesterase isof... 125 3e-29
TC14929 similar to UP|CAE76633 (CAE76633) Pectin methylesterase ... 122 2e-28
BP050971 119 1e-27
AV426815 111 4e-25
AU089161 63 5e-24
TC11594 similar to UP|Q9FXW9 (Q9FXW9) Pectin methylesterase-like... 100 1e-21
BF177669 99 2e-21
TC14589 similar to UP|Q9SI72 (Q9SI72) F23N19.14, partial (38%) 89 2e-18
>TC9337 similar to UP|PME_PRUPE (Q43062) Pectinesterase PPE8B precursor
(Pectin methylesterase) (PE) , partial (54%)
Length = 1145
Score = 232 bits (592), Expect(2) = 5e-98
Identities = 114/188 (60%), Positives = 141/188 (74%)
Frame = +2
Query: 302 GNGTVKTITEAIKKAPEHSRRRFIIYVRAGRYEENNLKVGKKKTNIMFIGDGRGKTVITG 361
G+G T+ EA+ AP++S +RF+IYV+ G YEE ++ KKK NIM +GDG TVI+G
Sbjct: 32 GSGNYTTVMEAVLAAPDYSMKRFVIYVKKGVYEEY-AEIKKKKWNIMMVGDGMDVTVISG 208
Query: 362 KRSVGDGMTTFHTASFAASGPGFMARDITFENYAGPEKHQAVALRVGSDHAVVYRCNIVG 421
RS GDG TTF +A+FA SG GF+AR ITF+N AGPEKHQAVALR SD AV +RC+I G
Sbjct: 209 NRSFGDGWTTFKSATFAVSGRGFIARGITFQNTAGPEKHQAVALRSDSDLAVFFRCSITG 388
Query: 422 YQDACYVHSNRQFFRECNIYGTVDFIFGNAAVVFQKCNIYARKPMAQQKNTITAQNRKDP 481
YQD+ Y H+ RQF++EC I GTVDFIFG+A VFQ CNI ARK + QKNTITAQ RKDP
Sbjct: 389 YQDSLYPHTMRQFYKECRITGTVDFIFGDATAVFQNCNIQARKGLQDQKNTITAQGRKDP 568
Query: 482 NQNTGISI 489
Q++G +
Sbjct: 569 GQSSGFFV 592
Score = 143 bits (360), Expect(2) = 5e-98
Identities = 69/120 (57%), Positives = 83/120 (68%)
Frame = +3
Query: 488 SIHDCRILPAPDLASSKGSIETYLGRPWKMYSRTVYMLSYMGDHVHPHGWLEWNGDFALK 547
S C I +L S GSI+TYLGRPWK YSRTV+M S+M D V GWLEWN F
Sbjct: 588 SFQYCNITADSELLPSVGSIQTYLGRPWKEYSRTVFMQSFMSDVVSSQGWLEWNTSFD-D 764
Query: 548 TLYYGEYMNFGPGAAIGQRVKWPGYRVITSTLEANRYTVAQFISGSSWLPSTGVAFLAGL 607
TL+YGEY+N+G GA +G RVKW GY V+ + EA ++TV QFI G+ WLPSTGVA+ GL
Sbjct: 765 TLFYGEYLNYGAGAGVGNRVKWGGYHVLNDS-EAGKFTVNQFIEGNLWLPSTGVAYTGGL 941
>TC17355 similar to UP|Q9FF77 (Q9FF77) Pectinesterase, partial (34%)
Length = 848
Score = 262 bits (669), Expect = 1e-70
Identities = 133/225 (59%), Positives = 162/225 (71%), Gaps = 3/225 (1%)
Frame = +1
Query: 295 IIVSKSGGNGTVKTITEAIKKAPEHSRRR---FIIYVRAGRYEENNLKVGKKKTNIMFIG 351
+IVS G +I +AI AP H++ + F+IYVR G YEE + V K+K NI+ +G
Sbjct: 154 VIVSPYNGTDNYTSIGDAIAAAPNHTKPQDGYFLIYVREGYYEEYVI-VPKEKKNILLVG 330
Query: 352 DGRGKTVITGKRSVGDGMTTFHTASFAASGPGFMARDITFENYAGPEKHQAVALRVGSDH 411
DG KT+ITG SV DG TTF++++FA SG F A D+TF N AGP KHQAVA+R +D
Sbjct: 331 DGINKTIITGNHSVIDGWTTFNSSTFAVSGERFTAVDVTFRNTAGPAKHQAVAVRNNADL 510
Query: 412 AVVYRCNIVGYQDACYVHSNRQFFRECNIYGTVDFIFGNAAVVFQKCNIYARKPMAQQKN 471
+ YRC+ GYQD YVHS RQF+REC+IYGTVDFIFGNAAVVFQ CNIYARKP+ QKN
Sbjct: 511 STFYRCSFEGYQDTLYVHSLRQFYRECDIYGTVDFIFGNAAVVFQNCNIYARKPLPFQKN 690
Query: 472 TITAQNRKDPNQNTGISIHDCRILPAPDLASSKGSIETYLGRPWK 516
+TAQ R DPNQNTGISI +C I APDLA+ S +YLGRPW+
Sbjct: 691 AVTAQGRTDPNQNTGISIQNCTIDAAPDLAAELNSTLSYLGRPWE 825
>TC9501 weakly similar to UP|Q9MBB6 (Q9MBB6) Pectin methylesterase ,
partial (29%)
Length = 934
Score = 256 bits (653), Expect = 1e-68
Identities = 126/256 (49%), Positives = 158/256 (61%), Gaps = 5/256 (1%)
Frame = +2
Query: 356 KTVITGKRSVGDGMTTFHTASFAASGPGFMARDITFENYAGPEKHQAVALRVGSDHAVVY 415
KT+ITG+++ DG+ T TA+FA PGF+A+ ITFEN AGP HQAVALR D +
Sbjct: 11 KTIITGRKNFADGVKTMQTATFANQAPGFIAKAITFENTAGPNGHQAVALRNQGDQSAFV 190
Query: 416 RCNIVGYQDACYVHSNRQFFRECNIYGTVDFIFGNAAVVFQKCNIYARKPMAQQKNTITA 475
C+I+GYQD YV +NRQF+R C I GT+DFIFG + + Q I RKP Q NT+TA
Sbjct: 191 GCHILGYQDTLYVQTNRQFYRNCVISGTIDFIFGTSPTLIQHSIIVVRKPNDNQFNTVTA 370
Query: 476 QNRKDPNQNTGISIHDCRILPAPDLASSKGSIETYLGRPWKMYSRTVYMLSYMGDHVHPH 535
N TGI I DC I+P L + I++YLGRPWK YSRTV M S +GD +HP
Sbjct: 371 DGTPTKNMPTGIVIQDCEIVPEAALFPVRFKIKSYLGRPWKEYSRTVVMESTLGDFIHPE 550
Query: 536 GWLEWNGDFALKTLYYGEYMNFGPGAAIGQRVKWPGYRVITSTLEANRYTVAQFI----- 590
GW W G FAL TLYY E+ N GPGA R+KW GYR + S EA ++T +F+
Sbjct: 551 GWFPWAGSFALDTLYYAEHANTGPGANTAGRIKWKGYRGVISRAEATQFTAGEFLKAGPR 730
Query: 591 SGSSWLPSTGVAFLAG 606
SG W+ + V G
Sbjct: 731 SGLDWIKALHVPHYLG 778
>CN825195
Length = 551
Score = 253 bits (645), Expect = 8e-68
Identities = 133/183 (72%), Positives = 154/183 (83%), Gaps = 1/183 (0%)
Frame = +2
Query: 204 LTNQDTCLEGFEDTSGTVKDQMVGNLKDLSELVSNSLAIFSASGDNDFTGVPIQNKRRLM 263
LTNQDTC +GF DTSG VKDQM NLKDLSELVSN LAIF+ +GD DF+GVPIQN RRL+
Sbjct: 17 LTNQDTCADGFADTSGDVKDQMAVNLKDLSELVSNCLAIFAGAGD-DFSGVPIQN-RRLL 190
Query: 264 GMSDISREFPKWLEKRDRRLLSLPVSEIQADIIVSKSGGNGTVKTITEAIKKAPEHSRRR 323
M D + FP+WL +RDRRLLSLPVS +QAD++VSK G NGT KTI EAIKK PE+ RR
Sbjct: 191 AMRDDN--FPRWLNRRDRRLLSLPVSAMQADVVVSKDG-NGTAKTIAEAIKKVPEYGSRR 361
Query: 324 FIIYVRAGRYEENNLKVGKKKTNIMFIGDGRGKTVITGKRSVGDG-MTTFHTASFAASGP 382
FIIYVRAGRYEENNLK+G+K+TN+M IGDG+GKTVITG ++V G +TTFHTASFAASG
Sbjct: 362 FIIYVRAGRYEENNLKIGRKRTNVMIIGDGKGKTVITGGKNVMYGLLTTFHTASFAASGA 541
Query: 383 GFM 385
GF+
Sbjct: 542 GFI 550
>TC10949 similar to UP|PME3_PHAVU (Q43111) Pectinesterase 3 precursor
(Pectin methylesterase 3) (PE 3) , partial (34%)
Length = 773
Score = 221 bits (562), Expect = 3e-58
Identities = 107/206 (51%), Positives = 135/206 (64%), Gaps = 1/206 (0%)
Frame = +3
Query: 399 KHQAVALRVGSDHAVVYRCNIVGYQDACYVHSNRQFFRECNIYGTVDFIFGNAAVVFQKC 458
KHQAVA+R GSD +V YRC+ G+QD Y HSNRQF+REC+I GT+DFIFGNAA +FQ C
Sbjct: 6 KHQAVAMRSGSDRSVFYRCSFDGFQDTLYAHSNRQFYRECDITGTIDFIFGNAASIFQNC 185
Query: 459 NIYARKPMAQQKNTITAQNRKDPNQNTGISIHDCRILPAPDLASSKGSIETYLGRPWKMY 518
I R+PM Q TITAQ +KDPNQNTGI I I P S + TYLGRPWK +
Sbjct: 186 KIMPRQPMPNQFYTITAQGKKDPNQNTGIVIQKSTITP----LESTYTAPTYLGRPWKDF 353
Query: 519 SRTVYMLSYMGDHVHPHGWLEWNGDF-ALKTLYYGEYMNFGPGAAIGQRVKWPGYRVITS 577
S T+ M S +G + P GW+ W + T++Y E+ N GPG+ + QRVKW GY+ +
Sbjct: 354 STTLIMQSEIGSFLKPVGWVSWVANVDPPATIFYAEHQNSGPGSDVAQRVKWTGYKSTLT 533
Query: 578 TLEANRYTVAQFISGSSWLPSTGVAF 603
+ ++T+A FI G WLP T VAF
Sbjct: 534 ADDLGKFTLASFIQGPEWLPDTAVAF 611
>TC16044 similar to UP|CAE76634 (CAE76634) Pectin methylesterase (Fragment)
, partial (43%)
Length = 564
Score = 194 bits (492), Expect = 5e-50
Identities = 87/110 (79%), Positives = 98/110 (89%)
Frame = +3
Query: 500 LASSKGSIETYLGRPWKMYSRTVYMLSYMGDHVHPHGWLEWNGDFALKTLYYGEYMNFGP 559
L +S S ETYLGRPWK+YSRTVYM+SY+GDHVH GWLEWN FAL TLYYGEYMN+GP
Sbjct: 3 LKASNDSFETYLGRPWKLYSRTVYMMSYIGDHVHQRGWLEWNTTFALDTLYYGEYMNYGP 182
Query: 560 GAAIGQRVKWPGYRVITSTLEANRYTVAQFISGSSWLPSTGVAFLAGLST 609
GAA+GQRV WPGYRVITS LEA+R+TVAQFI G++WLPSTGVAFL+GLST
Sbjct: 183 GAAVGQRVNWPGYRVITSALEASRFTVAQFILGATWLPSTGVAFLSGLST 332
>TC12620 weakly similar to UP|Q9FK05 (Q9FK05) Pectinesterase, partial (10%)
Length = 430
Score = 182 bits (461), Expect = 2e-46
Identities = 94/134 (70%), Positives = 115/134 (85%), Gaps = 5/134 (3%)
Frame = +3
Query: 9 SDPGGSSRRSTAPLTPSTT--SNSSPKSNKKLIMLSILAAVLIIASAISAALITVVRSRA 66
+DPGGSSRRSTAPLT STT S +S + KKL+ML++LAA+LI+AS +SA L+TV+RSR
Sbjct: 3 ADPGGSSRRSTAPLTGSTTVVSPNSSSTKKKLVMLAVLAAILIVASVVSAVLVTVIRSRG 182
Query: 67 SSN---NSNLLHSKPTQAISRTCSKTRYPSLCINSLLDFPGSTSASEQELVHISFNMTHR 123
S+ NSNL KPTQAISRTCS+TR+P+LCINSLLDFPGST+A+EQ+LVHISFNMTH
Sbjct: 183 GSSAGENSNL-RGKPTQAISRTCSRTRFPTLCINSLLDFPGSTTATEQDLVHISFNMTHH 359
Query: 124 HISKALFASSGLSY 137
HIS+AL+ SSGLS+
Sbjct: 360 HISRALYTSSGLSF 401
>TC8690 similar to UP|Q8RVX1 (Q8RVX1) Pectin methylesterase precursor
(Pectinesterase) , partial (59%)
Length = 1082
Score = 172 bits (435), Expect = 2e-43
Identities = 126/391 (32%), Positives = 193/391 (49%), Gaps = 14/391 (3%)
Frame = +3
Query: 13 GSSRRSTAPLTPSTTSNSS-----PKSNKKLIMLSILAAVLIIASAISAALITVVRSRAS 67
GSSR S+ T S KS K L + +I +SA+ A+ +
Sbjct: 45 GSSRASSQ*NRKMATQQQSLLDKPRKSFSKTFWLFLSLVAIISSSALIASYL-------K 203
Query: 68 SNNSNLLHSKPTQAISRTCSKTRYPSLCINSLLDFPGST-SASEQELVHISFNMTHR--- 123
+ NL S P C S C+ + + S SA++ ++I ++ +
Sbjct: 204 PTSFNLFLSPP-----HGCEHALDASSCLAHVSEVSQSPISATKDPKLNILISLMTKSTS 368
Query: 124 HISKALFASSGLSYTVANPRVRAAYEDCLELMDESMDAIRSSMDSLMTTSSTLSNDDGES 183
HI +A+ + + + NPR AA DC +LMD S+D + DS+M L+ D+ +S
Sbjct: 369 HIQEAMVKTKAIKNRINNPREEAALSDCEQLMDLSIDRV---WDSVMA----LTKDNTDS 527
Query: 184 RQFSNVAGSTEDVMTWLSAALTNQDTCLEGFEDTSGTVKDQMVGNLKDLSELVSNSLAIF 243
Q D WLS LTN TCL+G E S + M ++DL SLA+
Sbjct: 528 HQ---------DAHAWLSGVLTNHATCLDGLEGPSRAL---MEAEIEDLISRSKTSLALL 671
Query: 244 SA-----SGDNDFTGVPIQNKRRLMGMSDISREFPKWLEKRDRRLLSLPVSEIQADIIVS 298
+ G+ P+ +FP W+ ++DRRLL V ++ A+++V+
Sbjct: 672 VSVLAPKGGNEQIIDEPLDG------------DFPSWVTRKDRRLLESSVGDVNANVVVA 815
Query: 299 KSGGNGTVKTITEAIKKAPEHSRRRFIIYVRAGRYEENNLKVGKKKTNIMFIGDGRGKTV 358
K G +G KT+ EA+ AP+ + R++IYV+ G Y+EN +++GKKKTN+M GDG T+
Sbjct: 816 KDG-SGRFKTVAEAVASAPDSGKTRYVIYVKKGTYKEN-IEIGKKKTNVMLTGDGMDATI 989
Query: 359 ITGKRSVGDGMTTFHTASFAASGPGFMARDI 389
ITG +V DG TTF +A+ AA G GF+A+DI
Sbjct: 990 ITGNLNVIDGSTTFKSATVAAVGDGFIAQDI 1082
>TC10625 similar to UP|CAE76634 (CAE76634) Pectin methylesterase (Fragment)
, partial (32%)
Length = 538
Score = 92.4 bits (228), Expect(2) = 2e-36
Identities = 38/42 (90%), Positives = 39/42 (92%)
Frame = +2
Query: 528 MGDHVHPHGWLEWNGDFALKTLYYGEYMNFGPGAAIGQRVKW 569
M DHVHP GWLEWNGDFAL TLYYGEYMN+GPGAAIGQRVKW
Sbjct: 5 MSDHVHPRGWLEWNGDFALDTLYYGEYMNYGPGAAIGQRVKW 130
Score = 77.4 bits (189), Expect(2) = 2e-36
Identities = 36/46 (78%), Positives = 42/46 (91%)
Frame = +3
Query: 564 GQRVKWPGYRVITSTLEANRYTVAQFISGSSWLPSTGVAFLAGLST 609
G + PGYR+ITST+EA+R+TVAQFISGSSWLPSTGVA+LAGLST
Sbjct: 114 GNGLNGPGYRIITSTVEADRFTVAQFISGSSWLPSTGVAYLAGLST 251
>AI967495
Length = 390
Score = 141 bits (356), Expect = 3e-34
Identities = 69/122 (56%), Positives = 83/122 (67%), Gaps = 1/122 (0%)
Frame = +1
Query: 428 VHSNRQFFRECNIYGTVDFIFGNAAVVFQKCNIYARKPMAQQKNTITAQNRKDPNQNTGI 487
VH+ RQF+++C IYGTVD IFGNAAVVFQ C I+A+KP+ Q N ITAQ R DP QNTGI
Sbjct: 16 VHAQRQFYKQCQIYGTVDLIFGNAAVVFQNCKIFAKKPLDGQANMITAQGRDDPFQNTGI 195
Query: 488 SIHDCRILPAPDLASSKGSIETYLGRPWKMYSRTVYMLSYMGDHVHPHGWLEWNG-DFAL 546
+IH+ I APDL T LGRPW+ YSR V + ++M V P GW W+ DFAL
Sbjct: 196 TIHNSEIRAAPDLKPVVNKHNTSLGRPWQQYSRVVVLKTFMDTLVSPLGWSSWDDTDFAL 375
Query: 547 KT 548
T
Sbjct: 376 DT 381
>AV765593
Length = 501
Score = 141 bits (355), Expect = 3e-34
Identities = 67/121 (55%), Positives = 83/121 (68%)
Frame = -1
Query: 483 QNTGISIHDCRILPAPDLASSKGSIETYLGRPWKMYSRTVYMLSYMGDHVHPHGWLEWNG 542
Q+TG SIH+C I A DLASS G+ TYLGRPWK YSRTVYM + M + + GW W+G
Sbjct: 501 QDTGTSIHNCTIRAADDLASSNGATATYLGRPWKEYSRTVYMQTSMDNVIDSAGWRAWDG 322
Query: 543 DFALKTLYYGEYMNFGPGAAIGQRVKWPGYRVITSTLEANRYTVAQFISGSSWLPSTGVA 602
+FAL TLYY E+ N GPG+ RV W GY VI +T AN +TV+ F+ G WLP TGV+
Sbjct: 321 EFALSTLYYAEFNNSGPGSNTNGRVTWQGYHVINATDAAN-FTVSNFLLGDDWLPQTGVS 145
Query: 603 F 603
+
Sbjct: 144 Y 142
>TC9624 similar to UP|Q43234 (Q43234) Pectinmethylesterase precursor
(Pectinesterase) (Pectin methylesterase) (Fragment) ,
partial (33%)
Length = 586
Score = 127 bits (319), Expect = 5e-30
Identities = 59/106 (55%), Positives = 75/106 (70%), Gaps = 3/106 (2%)
Frame = +3
Query: 505 GSIETYLGRPWKMYSRTVYMLSYMGDHVHPHGWLEWNG---DFALKTLYYGEYMNFGPGA 561
GSI+TYLGRPWK YSRT+ + S + H+ P GW EW+ DF L+TLYYGEY+N G GA
Sbjct: 6 GSIKTYLGRPWKKYSRTIVLQSSIDSHIDPTGWAEWDAQSKDF-LQTLYYGEYLNSGAGA 182
Query: 562 AIGQRVKWPGYRVITSTLEANRYTVAQFISGSSWLPSTGVAFLAGL 607
G+RV WPG+ VI + EA+++TVAQ I G+ WL GV F+ GL
Sbjct: 183 GTGKRVNWPGFHVIKTAAEASKFTVAQLIQGNVWLKGKGVNFIEGL 320
>TC20170 similar to UP|Q9M5J0 (Q9M5J0) Pectin methylesterase isoform alpha
(Fragment) , partial (32%)
Length = 592
Score = 125 bits (313), Expect = 3e-29
Identities = 58/93 (62%), Positives = 68/93 (72%)
Frame = +1
Query: 516 KMYSRTVYMLSYMGDHVHPHGWLEWNGDFALKTLYYGEYMNFGPGAAIGQRVKWPGYRVI 575
K+YSRTV++ SYM D + P GWLEWN FAL TLYYGEY N GPG+ RVKW GYRVI
Sbjct: 1 KLYSRTVFLKSYMEDLIDPAGWLEWNDTFALDTLYYGEYQNRGPGSNPSARVKWGGYRVI 180
Query: 576 TSTLEANRYTVAQFISGSSWLPSTGVAFLAGLS 608
S+ EA+++TV QFI GS WL STG+ F LS
Sbjct: 181 NSSTEASQFTVGQFIQGSDWLNSTGIPFYFNLS 279
>TC14929 similar to UP|CAE76633 (CAE76633) Pectin methylesterase (Fragment)
, partial (24%)
Length = 693
Score = 122 bits (306), Expect = 2e-28
Identities = 59/104 (56%), Positives = 69/104 (65%)
Frame = +2
Query: 504 KGSIETYLGRPWKMYSRTVYMLSYMGDHVHPHGWLEWNGDFALKTLYYGEYMNFGPGAAI 563
K S TYLGRPWK YSRTV+M S + D + P GW EW+G+FAL TL Y EY GPGA
Sbjct: 11 KYSFPTYLGRPWKEYSRTVFMQSSISDVIDPAGWHEWSGNFALNTLVYREYQYPGPGAGT 190
Query: 564 GQRVKWPGYRVITSTLEANRYTVAQFISGSSWLPSTGVAFLAGL 607
+RV W G +VITS EA +T FI+GS+WL STG F GL
Sbjct: 191 SKRVAWKGVKVITSASEAQAFTPGSFIAGSNWLGSTGFPFSLGL 322
>BP050971
Length = 477
Score = 119 bits (299), Expect = 1e-27
Identities = 57/111 (51%), Positives = 73/111 (65%), Gaps = 1/111 (0%)
Frame = -1
Query: 498 PDLASSKGSIETYLGRPWKMYSRTVYMLSYMGDHVHPHGWLEWNGDFAL-KTLYYGEYMN 556
PD S GSI T+LGRPWK +SRTV+M S+M + +HP GWLEW L++GEY N
Sbjct: 474 PDFQPSNGSIPTFLGRPWKNFSRTVFMESFMSEMIHPEGWLEWETYTTYDDPLFFGEYPN 295
Query: 557 FGPGAAIGQRVKWPGYRVITSTLEANRYTVAQFISGSSWLPSTGVAFLAGL 607
GPG+ + RVKW GY V+ ++ A +TV FI G WLPS GV++ AGL
Sbjct: 294 KGPGSGVANRVKWRGYHVLNNS-PAMDFTVNPFIKGDLWLPSPGVSYHAGL 145
>AV426815
Length = 391
Score = 111 bits (277), Expect = 4e-25
Identities = 58/125 (46%), Positives = 86/125 (68%)
Frame = +3
Query: 260 RRLMGMSDISREFPKWLEKRDRRLLSLPVSEIQADIIVSKSGGNGTVKTITEAIKKAPEH 319
R+LM ++ +P+W+ DRRLL ++AD++V+ + G+G KT++ A+ APE
Sbjct: 36 RKLMEAGEV---WPEWISAADRRLLQ--AGTVKADVVVA-ADGSGNFKTVSAAVAAAPEK 197
Query: 320 SRRRFIIYVRAGRYEENNLKVGKKKTNIMFIGDGRGKTVITGKRSVGDGMTTFHTASFAA 379
S+ R++I ++AG Y EN ++V KKKTNIMF+GDGR T+ITG R+V DG TTFH+A+ A
Sbjct: 198 SKTRYVIKIKAGVYREN-VEVPKKKTNIMFLGDGRTTTIITGSRNVVDGSTTFHSATVAV 374
Query: 380 SGPGF 384
G F
Sbjct: 375 VGGNF 389
>AU089161
Length = 400
Score = 62.8 bits (151), Expect(3) = 5e-24
Identities = 33/66 (50%), Positives = 42/66 (63%), Gaps = 2/66 (3%)
Frame = +1
Query: 344 KTNIMF--IGDGRGKTVITGKRSVGDGMTTFHTASFAASGPGFMARDITFENYAGPEKHQ 401
+ N++F G G KTVITG ++ DG T +A+F P FMA+ + FEN AG KHQ
Sbjct: 58 RKNLIF*CTGXGPKKTVITGNKNFVDGYKTMRSATFTTVAPDFMAKSMGFENTAGASKHQ 237
Query: 402 AVALRV 407
AVALRV
Sbjct: 238 AVALRV 255
Score = 56.6 bits (135), Expect(3) = 5e-24
Identities = 23/45 (51%), Positives = 31/45 (68%)
Frame = +2
Query: 410 DHAVVYRCNIVGYQDACYVHSNRQFFRECNIYGTVDFIFGNAAVV 454
D + + C I GYQD Y H++RQF+R C I GTVDFIFG + ++
Sbjct: 263 DRSAFFDCAISGYQDTLYAHAHRQFYRNCEISGTVDFIFGYSTLL 397
Score = 28.9 bits (63), Expect(3) = 5e-24
Identities = 13/27 (48%), Positives = 20/27 (73%)
Frame = +3
Query: 325 IIYVRAGRYEENNLKVGKKKTNIMFIG 351
+IYV+AG Y+E +++ KKK NI+ G
Sbjct: 9 VIYVKAGVYDE-YIQIDKKKPNILMYG 86
>TC11594 similar to UP|Q9FXW9 (Q9FXW9) Pectin methylesterase-like protein,
partial (18%)
Length = 532
Score = 99.8 bits (247), Expect = 1e-21
Identities = 47/96 (48%), Positives = 60/96 (61%), Gaps = 4/96 (4%)
Frame = +3
Query: 508 ETYLGRPWKMYSRTVYMLSYMGDHVHPHGWLEWNGDFALKTLYYGEYMNFGPGAAIGQRV 567
+ YLGRPWK YSRTV++ S + V P GW+ WNG+FALKTLYYGE+ N GPG+ + RV
Sbjct: 63 KNYLGRPWKEYSRTVFIHSLLEALVTPQGWMPWNGEFALKTLYYGEFENSGPGSDLSLRV 242
Query: 568 KW----PGYRVITSTLEANRYTVAQFISGSSWLPST 599
W P V+T Y+ FI G W+PS+
Sbjct: 243 SWSSKVPAEHVLT-------YSAENFIQGDDWIPSS 329
>BF177669
Length = 336
Score = 99.4 bits (246), Expect = 2e-21
Identities = 49/101 (48%), Positives = 66/101 (64%)
Frame = +3
Query: 486 GISIHDCRILPAPDLASSKGSIETYLGRPWKMYSRTVYMLSYMGDHVHPHGWLEWNGDFA 545
GISI +C I P +L+ ++TYLGRPWK YS TV M S +G + P+GWL W G+ A
Sbjct: 42 GISIQNCSISPFGNLSY----VQTYLGRPWKNYSTTVVMQSTLGSFISPNGWLPWVGNSA 209
Query: 546 LKTLYYGEYMNFGPGAAIGQRVKWPGYRVITSTLEANRYTV 586
T++Y EY N G GA+ RVKW G + ITS +A+++TV
Sbjct: 210 PDTIFYAEYQNVGQGASTKNRVKWKGLKTITSK-QASKFTV 329
>TC14589 similar to UP|Q9SI72 (Q9SI72) F23N19.14, partial (38%)
Length = 826
Score = 89.4 bits (220), Expect = 2e-18
Identities = 66/222 (29%), Positives = 106/222 (47%), Gaps = 6/222 (2%)
Frame = +2
Query: 27 TSNSSPKSNKKLIMLSILAAVLIIASAISAALITVVRSRASSNNSNLLHSKPTQAISRTC 86
+ N S + + L++LS+L L++ +A SA A N N P + I +C
Sbjct: 38 SQNQSHMTGRVLLLLSLLVLSLVLFTAESAV--------ARGPNPN-----PIEFIKFSC 178
Query: 87 SKTRYPSLCINSLLDFPGSTSASEQEL----VHISFNMTHRHISKALFASSGLSYTVANP 142
TRYP++C+ +L + +EQ+L + +S +MT S A F P
Sbjct: 179 RATRYPAVCVQTLTRYAHVIRQNEQQLAITALTVSMSMTK---SSASFMKKMTKVKGIKP 349
Query: 143 RVRAAYEDCLELMDESMDAIRSSMDSLMTTSSTLSNDDGESRQFSNVAGSTEDVMTWLSA 202
R A +DC E MD S+D + S+ + T++ NV +V TW+SA
Sbjct: 350 REHGAVQDCKENMDNSVDRLNQSVKEMGLTAA------------GNVMWRMSNVQTWVSA 493
Query: 203 ALTNQDTCLEGF--EDTSGTVKDQMVGNLKDLSELVSNSLAI 242
ALT+Q+TCL+GF +K + + D S++ SN+LA+
Sbjct: 494 ALTDQNTCLDGFAHPQMDRNLKASIRARVVDASQVTSNALAL 619
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.317 0.131 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,781,966
Number of Sequences: 28460
Number of extensions: 131039
Number of successful extensions: 856
Number of sequences better than 10.0: 86
Number of HSP's better than 10.0 without gapping: 812
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 819
length of query: 609
length of database: 4,897,600
effective HSP length: 96
effective length of query: 513
effective length of database: 2,165,440
effective search space: 1110870720
effective search space used: 1110870720
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC134822.9


