

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC134822.10 - phase: 0
(333 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
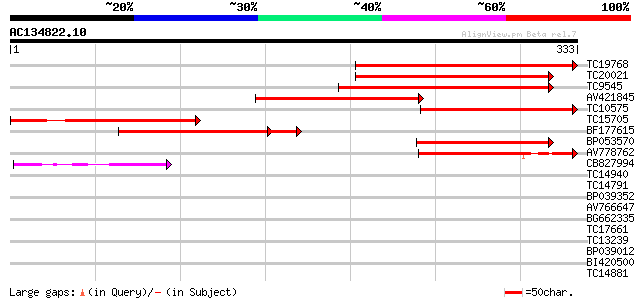
Score E
Sequences producing significant alignments: (bits) Value
TC19768 similar to UP|Q9FK06 (Q9FK06) Developmental protein SINA... 256 4e-69
TC20021 similar to UP|Q9LGX2 (Q9LGX2) ESTs AU082689(E31988), par... 227 2e-60
TC9545 similar to UP|Q9XGC3 (Q9XGC3) SINA2p, partial (40%) 224 2e-59
AV421845 214 1e-56
TC10575 homologue to UP|Q9LGX2 (Q9LGX2) ESTs AU082689(E31988), p... 191 2e-49
TC15705 similar to UP|Q9XGC2 (Q9XGC2) SINA1p, partial (27%) 190 3e-49
BF177615 154 3e-45
BP053570 113 4e-26
AV778762 113 4e-26
CB827994 83 6e-17
TC14940 34 0.039
TC14791 similar to UP|Q9LJ04 (Q9LJ04) ESTs AU082210(C53655), par... 33 0.067
BP039352 33 0.087
AV766647 32 0.15
BG662335 32 0.15
TC17661 weakly similar to UP|Q09085 (Q09085) Hydroxyproline-rich... 32 0.15
TC13239 homologue to UP|PHYB_SOYBN (P42499) Phytochrome B, parti... 32 0.19
BP039012 31 0.25
BI420500 31 0.33
TC14881 31 0.33
>TC19768 similar to UP|Q9FK06 (Q9FK06) Developmental protein SINA (Seven in
absentia), partial (47%)
Length = 572
Score = 256 bits (654), Expect = 4e-69
Identities = 119/130 (91%), Positives = 125/130 (95%)
Frame = +2
Query: 204 EVENATWMLTVFHCFGQYFCLHFEAFQLGMAPVYMAFLRFMGDENEARNYTYSLEVGANG 263
EVENATWMLTVFHCFGQYFCLHFEAFQLG APVYMAFLRFMGDE EARNY+YSLEVG NG
Sbjct: 5 EVENATWMLTVFHCFGQYFCLHFEAFQLGTAPVYMAFLRFMGDEREARNYSYSLEVGGNG 184
Query: 264 RKLIWEGTPRSIRDSHRKVRDSHDGLIIQRNMALFFSGGDRKELKLRVTGRIWKEQQNPD 323
RKL +EG+PRSIRDSH+KVRDSHDGLII RNMALFFSGGDRKELKLRVTGRIWKEQQNP+
Sbjct: 185 RKLTFEGSPRSIRDSHKKVRDSHDGLIIYRNMALFFSGGDRKELKLRVTGRIWKEQQNPE 364
Query: 324 GGVCIPNLCS 333
GGVC+PNLCS
Sbjct: 365 GGVCMPNLCS 394
>TC20021 similar to UP|Q9LGX2 (Q9LGX2) ESTs AU082689(E31988), partial (30%)
Length = 555
Score = 227 bits (578), Expect = 2e-60
Identities = 106/116 (91%), Positives = 111/116 (95%)
Frame = +2
Query: 204 EVENATWMLTVFHCFGQYFCLHFEAFQLGMAPVYMAFLRFMGDENEARNYTYSLEVGANG 263
EVENATWMLTVF CFGQYFCLHFEAFQLGMAPVY+AFLRFMGD+NEA+NYTYSLEVG NG
Sbjct: 2 EVENATWMLTVFSCFGQYFCLHFEAFQLGMAPVYIAFLRFMGDDNEAKNYTYSLEVGGNG 181
Query: 264 RKLIWEGTPRSIRDSHRKVRDSHDGLIIQRNMALFFSGGDRKELKLRVTGRIWKEQ 319
RK+IW+G PRSIRDSHR VRDS DGLIIQRNMALFFSGGDRKELKLRVTGRIWKEQ
Sbjct: 182 RKMIWQGVPRSIRDSHRMVRDSFDGLIIQRNMALFFSGGDRKELKLRVTGRIWKEQ 349
>TC9545 similar to UP|Q9XGC3 (Q9XGC3) SINA2p, partial (40%)
Length = 543
Score = 224 bits (571), Expect = 2e-59
Identities = 100/126 (79%), Positives = 118/126 (93%)
Frame = +3
Query: 194 NHRYVKSNPREVENATWMLTVFHCFGQYFCLHFEAFQLGMAPVYMAFLRFMGDENEARNY 253
NHRYVKSNP EVENATWMLT+F+ FG++FCLHFEAFQ+G APVYMAFLRF+G+++EA+ +
Sbjct: 15 NHRYVKSNPHEVENATWMLTIFNSFGRHFCLHFEAFQIGTAPVYMAFLRFLGEDSEAKKF 194
Query: 254 TYSLEVGANGRKLIWEGTPRSIRDSHRKVRDSHDGLIIQRNMALFFSGGDRKELKLRVTG 313
+YSLEVGANGRKL W+G PRSIRDSHRKVRDS DGLIIQRN+ L+FSGGDR+ELKLR+TG
Sbjct: 195 SYSLEVGANGRKLTWQGIPRSIRDSHRKVRDSQDGLIIQRNLGLYFSGGDRQELKLRITG 374
Query: 314 RIWKEQ 319
RIWKE+
Sbjct: 375 RIWKEE 392
>AV421845
Length = 300
Score = 214 bits (546), Expect = 1e-56
Identities = 96/99 (96%), Positives = 97/99 (97%)
Frame = +2
Query: 145 KLKHETICNFRPYTCPYAGSECSAVGDINFLVAHLRDDHKVDMHTGCTFNHRYVKSNPRE 204
KLKHETICNFRPY+CPYAGSECS VGDI FLVAHLRDDHKVDMHTGCTFNHRYVKSNPRE
Sbjct: 2 KLKHETICNFRPYSCPYAGSECSVVGDIPFLVAHLRDDHKVDMHTGCTFNHRYVKSNPRE 181
Query: 205 VENATWMLTVFHCFGQYFCLHFEAFQLGMAPVYMAFLRF 243
VENATWMLTVFHCFGQYFCLHFEAFQLGMAPVYMAFLRF
Sbjct: 182 VENATWMLTVFHCFGQYFCLHFEAFQLGMAPVYMAFLRF 298
>TC10575 homologue to UP|Q9LGX2 (Q9LGX2) ESTs AU082689(E31988), partial
(24%)
Length = 602
Score = 191 bits (484), Expect = 2e-49
Identities = 89/92 (96%), Positives = 91/92 (98%)
Frame = +2
Query: 242 RFMGDENEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRKVRDSHDGLIIQRNMALFFSG 301
RFMGDENEARNY+YSLEVGANGRKLIWEGTPRS+RDSHRKVRDSHDGLIIQRNMALFFSG
Sbjct: 2 RFMGDENEARNYSYSLEVGANGRKLIWEGTPRSVRDSHRKVRDSHDGLIIQRNMALFFSG 181
Query: 302 GDRKELKLRVTGRIWKEQQNPDGGVCIPNLCS 333
GDRKELKLRVTGRIWKEQQNPD GVCIPNLCS
Sbjct: 182 GDRKELKLRVTGRIWKEQQNPDAGVCIPNLCS 277
>TC15705 similar to UP|Q9XGC2 (Q9XGC2) SINA1p, partial (27%)
Length = 674
Score = 190 bits (482), Expect = 3e-49
Identities = 91/113 (80%), Positives = 94/113 (82%), Gaps = 1/113 (0%)
Frame = +1
Query: 1 MDLDSIECVSSSDGMDEDEIQHRILHPHHQQHHHHSEFSSLKPRSGG-NNNHGVIGSTAI 59
MDLDSIECVSSSDGMDEDEI HHHSEFSS K R+GG NNN+ +G TAI
Sbjct: 364 MDLDSIECVSSSDGMDEDEIHA----------HHHSEFSSTKARNGGGNNNNAAMGPTAI 513
Query: 60 APATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGD 112
PATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGD
Sbjct: 514 TPATSVHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGD 672
>BF177615
Length = 347
Score = 154 bits (390), Expect(2) = 3e-45
Identities = 68/90 (75%), Positives = 76/90 (83%)
Frame = +1
Query: 65 VHELLECPVCTNSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQELGDIRCLALEKVAES 124
V++LL+CPVC N MYPPIHQC NGHTLCS CK VHN CPTC +L +IRCLALEKVAES
Sbjct: 25 VYDLLKCPVCKNLMYPPIHQCPNGHTLCSNCKIEVHNICPTCDHDLENIRCLALEKVAES 204
Query: 125 LELPCKYYSLGCPEIFPYYSKLKHETICNF 154
LELPCKY SLGC +IFPYY+KLKHE I +F
Sbjct: 205 LELPCKYQSLGCHDIFPYYTKLKHEQIVDF 294
Score = 43.9 bits (102), Expect(2) = 3e-45
Identities = 16/20 (80%), Positives = 17/20 (85%)
Frame = +3
Query: 152 CNFRPYTCPYAGSECSAVGD 171
C FRPY CPYAGSECS +GD
Sbjct: 285 CGFRPYNCPYAGSECSVMGD 344
>BP053570
Length = 463
Score = 113 bits (283), Expect = 4e-26
Identities = 50/80 (62%), Positives = 65/80 (80%)
Frame = -2
Query: 240 FLRFMGDENEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRKVRDSHDGLIIQRNMALFF 299
FLRF+G+++EA + ++LEVGAN RKLIW G PRSIR SHRKVRD DGLII R++AL+F
Sbjct: 462 FLRFLGEDHEASKFRFTLEVGANSRKLIWPGIPRSIRHSHRKVRDCQDGLIIPRHLALYF 283
Query: 300 SGGDRKELKLRVTGRIWKEQ 319
S GD+ LK ++TG IWK++
Sbjct: 282 SSGDKPPLKFKITGHIWKDE 223
>AV778762
Length = 512
Score = 113 bits (283), Expect = 4e-26
Identities = 61/95 (64%), Positives = 74/95 (77%), Gaps = 2/95 (2%)
Frame = -3
Query: 241 LRFMGDENEARNYTYSLEVGANGRKLIWEGTPRSIRDSHRKVRDSHDGLIIQRNMALFFS 300
LRFMGDE EAR+Y+YSLEVG +GRKL +EG+PRSIRDSH++VRDSHDGL++ RNMA F
Sbjct: 510 LRFMGDEREARSYSYSLEVGGSGRKLTYEGSPRSIRDSHKQVRDSHDGLVVHRNMAHFLL 331
Query: 301 --GGDRKELKLRVTGRIWKEQQNPDGGVCIPNLCS 333
G R E VTG+IWK ++ P GGV IPN+ S
Sbjct: 330 RWG**RVE----VTGKIWK-RRTPVGGVDIPNMYS 241
Score = 30.0 bits (66), Expect = 0.57
Identities = 18/40 (45%), Positives = 20/40 (50%), Gaps = 4/40 (10%)
Frame = -1
Query: 297 LFFSGGDRKELKLRV----TGRIWKEQQNPDGGVCIPNLC 332
+FFSGGD KELKLR +W E P CI C
Sbjct: 341 IFFSGGDSKELKLRERYGNDEPLWVEWTYP---TCIARFC 231
>CB827994
Length = 545
Score = 83.2 bits (204), Expect = 6e-17
Identities = 48/94 (51%), Positives = 54/94 (57%), Gaps = 1/94 (1%)
Frame = +1
Query: 3 LDSIECVSSSDGMDEDEIQHRILHPHHQQHHHHSEFSSLKPRSGGNNNHGVIGSTAIAPA 62
++SI S+ M+EDE HPH +FSS I PA
Sbjct: 340 MESISIDSTVTMMEEDE------HPH--------QFSSTSKLHN------------IGPA 441
Query: 63 T-SVHELLECPVCTNSMYPPIHQCHNGHTLCSTC 95
T SVH+LLECPVCTNSMYPPIHQCHNGHTLCSTC
Sbjct: 442 TTSVHDLLECPVCTNSMYPPIHQCHNGHTLCSTC 543
>TC14940
Length = 781
Score = 33.9 bits (76), Expect = 0.039
Identities = 18/74 (24%), Positives = 31/74 (41%), Gaps = 4/74 (5%)
Frame = +2
Query: 61 PATSVHELLECPVCTNSMYPPIH-QCHNGHTLCSTC---KTRVHNRCPTCRQELGDIRCL 116
P + CP+C + + +C GH C C N+CPTCR+++ +
Sbjct: 119 PEPPKEPVFSCPICMGPLVEEMSTRC--GHIFCKACIKAAISAQNKCPTCRKKVTVKELI 292
Query: 117 ALEKVAESLELPCK 130
+ + + PCK
Sbjct: 293 RVFLPSTN*RFPCK 334
>TC14791 similar to UP|Q9LJ04 (Q9LJ04) ESTs AU082210(C53655), partial (37%)
Length = 1779
Score = 33.1 bits (74), Expect = 0.067
Identities = 19/60 (31%), Positives = 30/60 (49%), Gaps = 2/60 (3%)
Frame = +2
Query: 5 SIECVSSSDGMDEDEIQHRILHPHHQQHH--HHSEFSSLKPRSGGNNNHGVIGSTAIAPA 62
+++ S G +++ QH+ H HH HH H+ K SGG +HG G +AP+
Sbjct: 1121 AVDGPKGSKGYHQNQHQHQ--HQHHSHHHQPHYQPHKKHKYNSGGGPSHGG-GGHLMAPS 1291
>BP039352
Length = 528
Score = 32.7 bits (73), Expect = 0.087
Identities = 11/13 (84%), Positives = 12/13 (91%)
Frame = +2
Query: 24 ILHPHHQQHHHHS 36
+LHPHHQQHHH S
Sbjct: 386 LLHPHHQQHHHPS 424
>AV766647
Length = 349
Score = 32.0 bits (71), Expect = 0.15
Identities = 10/15 (66%), Positives = 11/15 (72%)
Frame = -3
Query: 21 QHRILHPHHQQHHHH 35
QH+ HPHH HHHH
Sbjct: 344 QHQHPHPHHHHHHHH 300
Score = 27.3 bits (59), Expect = 3.7
Identities = 9/15 (60%), Positives = 9/15 (60%)
Frame = -3
Query: 21 QHRILHPHHQQHHHH 35
QH H HH HHHH
Sbjct: 338 QHPHPHHHHHHHHHH 294
Score = 26.6 bits (57), Expect = 6.3
Identities = 8/14 (57%), Positives = 9/14 (64%)
Frame = -3
Query: 22 HRILHPHHQQHHHH 35
H+ HPH HHHH
Sbjct: 347 HQHQHPHPHHHHHH 306
>BG662335
Length = 336
Score = 32.0 bits (71), Expect = 0.15
Identities = 10/15 (66%), Positives = 11/15 (72%)
Frame = +1
Query: 21 QHRILHPHHQQHHHH 35
QH+ HPHH HHHH
Sbjct: 286 QHQHPHPHHHHHHHH 330
Score = 27.7 bits (60), Expect = 2.8
Identities = 11/32 (34%), Positives = 14/32 (43%)
Frame = +1
Query: 4 DSIECVSSSDGMDEDEIQHRILHPHHQQHHHH 35
D+ + G + QH H HH HHHH
Sbjct: 241 DAYDMRKQLKGKQLHQHQHPHPHHHHHHHHHH 336
>TC17661 weakly similar to UP|Q09085 (Q09085) Hydroxyproline-rich
glycoprotein (HRGP) (Fragment), partial (8%)
Length = 417
Score = 32.0 bits (71), Expect = 0.15
Identities = 21/68 (30%), Positives = 28/68 (40%), Gaps = 9/68 (13%)
Frame = +2
Query: 26 HPHHQQHHHHS-EFSSLKP--------RSGGNNNHGVIGSTAIAPATSVHELLECPVCTN 76
H HH HHHH+ F+ L P + H I ++ P T+ LL P T
Sbjct: 32 HRHHHHHHHHAILFTHLHPLQLLHHTHMDHRESTHSTIPHSSTHPTTN---LLHLPPQTA 202
Query: 77 SMYPPIHQ 84
P+HQ
Sbjct: 203PFQDPLHQ 226
>TC13239 homologue to UP|PHYB_SOYBN (P42499) Phytochrome B, partial (12%)
Length = 441
Score = 31.6 bits (70), Expect = 0.19
Identities = 26/95 (27%), Positives = 39/95 (40%), Gaps = 5/95 (5%)
Frame = -2
Query: 21 QHRILHPHHQQHHHHSEFSSLKPRSGGNNNHGVIGSTAI-----APATSVHELLECPVCT 75
+H LH Q H E +S+ R + GV+ + APAT+ + ++ P+
Sbjct: 302 RHLKLHVQLQPKSLHEELTSIPQRKRDTPSRGVVTNNKPPQPHRAPATNPNRVIIIPIDD 123
Query: 76 NSMYPPIHQCHNGHTLCSTCKTRVHNRCPTCRQEL 110
N + + H GH L S R R P Q L
Sbjct: 122 NRHHQRRDRTHVGHVL-SMATVRSTERGPHQTQRL 21
>BP039012
Length = 582
Score = 31.2 bits (69), Expect = 0.25
Identities = 11/22 (50%), Positives = 14/22 (63%)
Frame = +2
Query: 24 ILHPHHQQHHHHSEFSSLKPRS 45
+L PHH QHHHH++ P S
Sbjct: 230 VLSPHHPQHHHHTK*QYYTPSS 295
>BI420500
Length = 539
Score = 30.8 bits (68), Expect = 0.33
Identities = 20/89 (22%), Positives = 38/89 (42%)
Frame = -1
Query: 143 YSKLKHETICNFRPYTCPYAGSECSAVGDINFLVAHLRDDHKVDMHTGCTFNHRYVKSNP 202
+ K + + NF CP C V + FL+ + + + H Y+K +P
Sbjct: 284 FLKEEQRSAQNFHQNDCPLTTLNCMTVLTMLFLLQTSSNS*QRNYHYLLNLLLHYLKDDP 105
Query: 203 REVENATWMLTVFHCFGQYFCLHFEAFQL 231
+ + ++L +F F +++ F FQL
Sbjct: 104 KISDQKGFLLGLFGWFEEHYHAAFAHFQL 18
>TC14881
Length = 609
Score = 30.8 bits (68), Expect = 0.33
Identities = 19/68 (27%), Positives = 29/68 (41%), Gaps = 8/68 (11%)
Frame = +3
Query: 54 IGSTAIAPATSVHELLECPVCTN-----SMYPPIHQCHNGHTLCSTCKTRVH---NRCPT 105
IGS+ A EC +C ++ P H C +CS C + N+CP
Sbjct: 48 IGSSTAADFDDNDPQKECVICMTEPKDTAVLPCRHMC-----MCSDCAKELRLQSNKCPI 212
Query: 106 CRQELGDI 113
CRQ + ++
Sbjct: 213 CRQPIEEL 236
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.323 0.138 0.452
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,461,742
Number of Sequences: 28460
Number of extensions: 159112
Number of successful extensions: 2316
Number of sequences better than 10.0: 161
Number of HSP's better than 10.0 without gapping: 1735
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2078
length of query: 333
length of database: 4,897,600
effective HSP length: 91
effective length of query: 242
effective length of database: 2,307,740
effective search space: 558473080
effective search space used: 558473080
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 55 (25.8 bits)
Medicago: description of AC134822.10


