

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC134322.21 - phase: 0
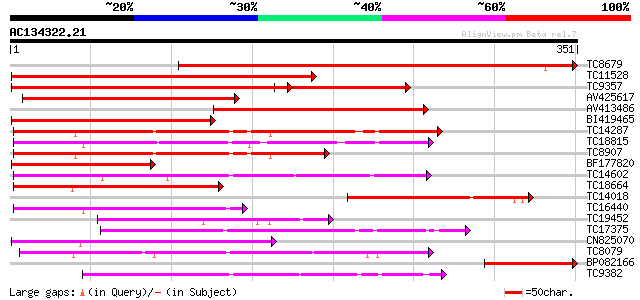
(351 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC8679 homologue to UP|Q43466 (Q43466) Protein kinase 3 , partia... 412 e-116
TC11528 homologue to UP|Q39868 (Q39868) Protein kinase , partial... 344 1e-95
TC9357 homologue to UP|Q39193 (Q39193) Protein kinase (Protein k... 314 1e-86
AV425617 259 5e-70
AV413486 223 5e-59
BI419465 219 4e-58
TC14287 similar to UP|Q9LEU7 (Q9LEU7) Serine/threonine protein k... 196 4e-51
TC18815 similar to UP|Q8W1D5 (Q8W1D5) CBL-interacting protein ki... 189 8e-49
TC8907 similar to UP|Q8H0X3 (Q8H0X3) Serine/threonine protein ki... 162 8e-41
BF177820 154 3e-38
TC14602 UP|Q8GS56 (Q8GS56) Phosphoenolpyruvate carboxylase kinas... 135 1e-32
TC18664 similar to UP|Q944I6 (Q944I6) At2g30360/T9D9.17, partial... 122 1e-28
TC14018 similar to UP|Q43465 (Q43465) Protein kinase 2, partial ... 118 2e-27
TC16440 similar to UP|Q93Y18 (Q93Y18) SNF1 related protein kinas... 105 1e-23
TC19452 homologue to UP|Q8S9J9 (Q8S9J9) At1g14000/F7A19_9, parti... 94 3e-20
TC17375 mitogen-activated kinase kinase kinase alpha [Lotus corn... 93 6e-20
CN825070 92 1e-19
TC8079 homologue to UP|Q9M6S1 (Q9M6S1) MAP kinase 3, complete 92 2e-19
BP082166 82 1e-16
TC9382 weakly similar to UP|ABL_DROME (P00522) Tyrosine-protein ... 82 1e-16
>TC8679 homologue to UP|Q43466 (Q43466) Protein kinase 3 , partial (70%)
Length = 972
Score = 412 bits (1058), Expect = e-116
Identities = 202/251 (80%), Positives = 222/251 (87%), Gaps = 4/251 (1%)
Frame = +2
Query: 105 QQLISGVSYCHSMQICHRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPA 164
+QLISGV YCH++QICHRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPA
Sbjct: 2 EQLISGVHYCHALQICHRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPA 181
Query: 165 YIAPEVLSRREYDGKLADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIP 224
YIAPEVLSRREYDGKLADVWSCGVTLYVMLVGAYPFED DDP+NFRKTI RIMAVQYKIP
Sbjct: 182 YIAPEVLSRREYDGKLADVWSCGVTLYVMLVGAYPFEDHDDPRNFRKTIQRIMAVQYKIP 361
Query: 225 DYVHISQECRHLLSRIFVASPARRITIKEIKSHPWFLKNLPRELTEMAQAVYY-RKENPT 283
DYVHISQ+CRHLLSRIFVA+P RRI++KEIK+HPWFLKNLPRELTE AQAVYY R N +
Sbjct: 362 DYVHISQDCRHLLSRIFVANPLRRISLKEIKNHPWFLKNLPRELTESAQAVYYQRGGNLS 541
Query: 284 YSLQSIEDIMKIVEEAKNPPQASRSVGGFGWGGEEDDDEINEAEAEL---EEDEYEKRVK 340
+S+QS+E+IMKIV EA+ PP SR V GFGW GEE+D+E E E E EEDEY+KRVK
Sbjct: 542 FSVQSVEEIMKIVGEAREPPPVSRPVKGFGWEGEEEDEEDVEEEVEEEEDEEDEYDKRVK 721
Query: 341 EAQESGEIHVN 351
E SGE ++
Sbjct: 722 EVHASGEFQIS 754
>TC11528 homologue to UP|Q39868 (Q39868) Protein kinase , partial (56%)
Length = 640
Score = 344 bits (883), Expect = 1e-95
Identities = 163/189 (86%), Positives = 182/189 (96%)
Frame = +3
Query: 2 EKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVAREIINHRSLRHPNII 61
E+YE +K++GSGNFGVARL R K+T +LVA+KYIERG KIDENV REIINHRSLRHPNII
Sbjct: 72 ERYEPLKELGSGNFGVARLARDKNTGELVAVKYIERGKKIDENVQREIINHRSLRHPNII 251
Query: 62 RFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQICH 121
RFKEV+LTPTHL IV+EYA+GGELF+RICSAGRFSEDEARYFFQQLISGVSYCHSM+ICH
Sbjct: 252 RFKEVLLTPTHLAIVLEYASGGELFERICSAGRFSEDEARYFFQQLISGVSYCHSMEICH 431
Query: 122 RDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKLA 181
RDLKLENTLLDG+P+PRLKICDFGYSKS++LHS+PKSTVGTPAYIAPEVLSR+EYDGK+A
Sbjct: 432 RDLKLENTLLDGNPSPRLKICDFGYSKSAILHSQPKSTVGTPAYIAPEVLSRKEYDGKVA 611
Query: 182 DVWSCGVTL 190
DVWSCGVTL
Sbjct: 612 DVWSCGVTL 638
>TC9357 homologue to UP|Q39193 (Q39193) Protein kinase (Protein kinase,
41K) , partial (71%)
Length = 1181
Score = 314 bits (805), Expect = 1e-86
Identities = 147/174 (84%), Positives = 168/174 (96%)
Frame = +1
Query: 2 EKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVAREIINHRSLRHPNII 61
++Y++V+DIGSGNFGVARLM+ K TK+LVA+KYIERG KIDENV REIINHRSLRHPNI+
Sbjct: 355 DRYDLVRDIGSGNFGVARLMQDKQTKELVAVKYIERGDKIDENVKREIINHRSLRHPNIV 534
Query: 62 RFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQICH 121
RFKEV+LTPTHL IVMEYA+GGELF+RIC+AGRF+EDEAR+FFQQLISGVSYCH+MQ+CH
Sbjct: 535 RFKEVILTPTHLAIVMEYASGGELFERICNAGRFTEDEARFFFQQLISGVSYCHAMQVCH 714
Query: 122 RDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRRE 175
RDLKLENTLLDGSP PRLKICDFGYSKSS+LHS+PKSTVGTPAYIAPEVLS++E
Sbjct: 715 RDLKLENTLLDGSPTPRLKICDFGYSKSSVLHSQPKSTVGTPAYIAPEVLSKQE 876
Score = 108 bits (271), Expect = 1e-24
Identities = 51/84 (60%), Positives = 63/84 (74%)
Frame = +3
Query: 165 YIAPEVLSRREYDGKLADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIP 224
Y + ++ R +L SCGVTLYVMLVGAYPFED ++PK+FRKTI R+++VQY IP
Sbjct: 846 YCSRSIIETRVMMARLQMSGSCGVTLYVMLVGAYPFEDPNEPKDFRKTIQRVLSVQYSIP 1025
Query: 225 DYVHISQECRHLLSRIFVASPARR 248
D+V IS ECRHL+SRIFV PA R
Sbjct: 1026DFVQISPECRHLISRIFVFDPAER 1097
>AV425617
Length = 405
Score = 259 bits (662), Expect = 5e-70
Identities = 125/134 (93%), Positives = 131/134 (97%)
Frame = +2
Query: 9 DIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVAREIINHRSLRHPNIIRFKEVVL 68
DIGSGNFGVARLMRHK+TK+LVAMKYIERG KIDENVAREIINHRSLRHPNIIRFKE+VL
Sbjct: 2 DIGSGNFGVARLMRHKETKELVAMKYIERGLKIDENVAREIINHRSLRHPNIIRFKELVL 181
Query: 69 TPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQICHRDLKLEN 128
TPTHL IVMEYAAGGELF+RIC+AGRFSEDEARYFFQQLISGV YCH+MQICHRDLKLEN
Sbjct: 182 TPTHLAIVMEYAAGGELFERICTAGRFSEDEARYFFQQLISGVCYCHAMQICHRDLKLEN 361
Query: 129 TLLDGSPAPRLKIC 142
TLLDGSPAPRLKIC
Sbjct: 362 TLLDGSPAPRLKIC 403
>AV413486
Length = 402
Score = 223 bits (567), Expect = 5e-59
Identities = 104/133 (78%), Positives = 116/133 (87%)
Frame = +3
Query: 127 ENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDGKLADVWSC 186
ENTLLDGSPA LKICDFGYSKSS+LHS+PKSTVGTPAYIAPEVL ++EYDGK+ADVWSC
Sbjct: 3 ENTLLDGSPALHLKICDFGYSKSSVLHSQPKSTVGTPAYIAPEVLLKQEYDGKIADVWSC 182
Query: 187 GVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHLLSRIFVASPA 246
GVTLYVMLVG YPFED DDPK+FRKTI R++ VQY IPD+V +S ECRHL+SRIFV PA
Sbjct: 183 GVTLYVMLVGTYPFEDPDDPKDFRKTIQRVLTVQYSIPDFVQVSPECRHLISRIFVFDPA 362
Query: 247 RRITIKEIKSHPW 259
RITI EI + W
Sbjct: 363 ERITIPEILKNEW 401
>BI419465
Length = 487
Score = 219 bits (559), Expect = 4e-58
Identities = 105/126 (83%), Positives = 118/126 (93%)
Frame = +3
Query: 2 EKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVAREIINHRSLRHPNII 61
++Y+ V+DIGSGNFGVARLMR K TK+LVA+KYIERG KI ENV REIINHRSLRHPNI+
Sbjct: 108 DRYDFVRDIGSGNFGVARLMRDKHTKELVAVKYIERGDKIYENVKREIINHRSLRHPNIV 287
Query: 62 RFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQICH 121
RFKEV+LTPTHL IVMEYA+GGELF++I SAGRF+EDEAR+FFQQLISGVSYCHSMQICH
Sbjct: 288 RFKEVILTPTHLAIVMEYASGGELFEKIHSAGRFTEDEARFFFQQLISGVSYCHSMQICH 467
Query: 122 RDLKLE 127
RDLKLE
Sbjct: 468 RDLKLE 485
>TC14287 similar to UP|Q9LEU7 (Q9LEU7) Serine/threonine protein kinase-like
protein (CBL-interacting protein kinase 5), partial
(75%)
Length = 1999
Score = 196 bits (499), Expect = 4e-51
Identities = 113/275 (41%), Positives = 166/275 (60%), Gaps = 9/275 (3%)
Frame = +3
Query: 3 KYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGH----KIDENVAREIINHRSLRHP 58
KYE+ + +G GNF R+ T + VA+K I++ ++ + + RE+ R +RHP
Sbjct: 429 KYEMGRVLGQGNFAKVYHGRNLATNENVAIKVIKKEKLKKDRLVKQIKREVSVMRLVRHP 608
Query: 59 NIIRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQ 118
+I+ KEV+ T + +VMEY GGELF ++ + G+ +ED+AR +FQQLIS V +CHS
Sbjct: 609 HIVELKEVMATKGKIFLVMEYVKGGELFTKV-NKGKLNEDDARKYFQQLISAVDFCHSRG 785
Query: 119 ICHRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTV-----GTPAYIAPEVLSR 173
+ HRDLK EN LLD + LK+ DFG S +L R + GTPAY+APEVL +
Sbjct: 786 VTHRDLKPENLLLDENED--LKVSDFGLS--ALPEQRRDDGMLVTPCGTPAYVAPEVLKK 953
Query: 174 REYDGKLADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQEC 233
+ YDG AD+WSCGV LY +L G PF+ ++ + +RK +Y+ P++ IS +
Sbjct: 954 KGYDGSKADIWSCGVILYALLSGYLPFQGENVMRIYRKAFK----AEYEFPEW--ISPQA 1115
Query: 234 RHLLSRIFVASPARRITIKEIKSHPWFLKNLPREL 268
++L+S + VA P +R +I EI S PWF R L
Sbjct: 1116KNLISNLLVADPEKRYSIPEIISDPWFQYGFMRPL 1220
>TC18815 similar to UP|Q8W1D5 (Q8W1D5) CBL-interacting protein kinase
CIPK25, partial (58%)
Length = 979
Score = 189 bits (479), Expect = 8e-49
Identities = 110/271 (40%), Positives = 152/271 (55%), Gaps = 11/271 (4%)
Frame = +2
Query: 3 KYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDEN----VAREIINHRSLRHP 58
KYE+ + +G G + + + VA+K + + E + REI R +RHP
Sbjct: 35 KYEMGRVLGKGTLAKVYFAKEITSGEGVAIKVMSKARIKKEGMMDQIKREISIMRLVRHP 214
Query: 59 NIIRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQ 118
NI+ KEV+ T T + +MEY GGELF ++ G+ +D AR +FQQLIS V YCHS
Sbjct: 215 NIVNLKEVMATKTKIFFIMEYIRGGELFAKVAK-GKLKDDLARRYFQQLISAVDYCHSRG 391
Query: 119 ICHRDLKLENTLLDGSPAPRLKICDFGYS-------KSSLLHSRPKSTVGTPAYIAPEVL 171
+ HRDLK EN LLD + LK+ DFG S + LLH++ GTPAY+APEVL
Sbjct: 392 VSHRDLKPENLLLDENE--NLKVSDFGLSGLPEQLRQDGLLHTQ----CGTPAYVAPEVL 553
Query: 172 SRREYDGKLADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQ 231
++ YDG D WSCGV LY +L G PF+ + N N+++ +++ P + S
Sbjct: 554 RKKGYDGFKTDTWSCGVILYALLAGCLPFQHE----NLMTMYNKVLRAEFQFPPW--FSP 715
Query: 232 ECRHLLSRIFVASPARRITIKEIKSHPWFLK 262
E + L+S+I VA P RRITI I WF K
Sbjct: 716 ESKKLISKILVADPNRRITISSIMRVSWFQK 808
>TC8907 similar to UP|Q8H0X3 (Q8H0X3) Serine/threonine protein kinase-like
protein, partial (44%)
Length = 941
Score = 162 bits (410), Expect = 8e-41
Identities = 91/205 (44%), Positives = 128/205 (62%), Gaps = 9/205 (4%)
Frame = +3
Query: 3 KYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGH----KIDENVAREIINHRSLRHP 58
KYE+ K +G GNF R+ +T + VA+K I++ ++ + + RE+ R +RHP
Sbjct: 339 KYEIGKILGQGNFAKVYHGRNMETNESVAIKVIKKERLKKERLVKQIKREVSVMRLVRHP 518
Query: 59 NIIRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQ 118
+I+ KEV+ T T + +V+EY GGELF ++ + G+ +E AR +FQQLIS V +CHS
Sbjct: 519 HIVELKEVMATKTKIFMVVEYVKGGELFAKL-TKGKMTEVAARKYFQQLISAVDFCHSRG 695
Query: 119 ICHRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTV-----GTPAYIAPEVLSR 173
+ HRDLK EN LLD + LK+ DFG S SL R + GTPAY+APEVL +
Sbjct: 696 VTHRDLKPENLLLDDNED--LKVSDFGLS--SLPEQRRSDGMLLTPCGTPAYVAPEVLKK 863
Query: 174 REYDGKLADVWSCGVTLYVMLVGAY 198
+ YDG AD+WSCGV LY +L G +
Sbjct: 864 KGYDGSKADIWSCGVILYALLCGIF 938
>BF177820
Length = 509
Score = 154 bits (388), Expect = 3e-38
Identities = 72/89 (80%), Positives = 83/89 (92%)
Frame = +1
Query: 2 EKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVAREIINHRSLRHPNII 61
++YE+V+DIGSGNFGVARLM+ K T +LVA+KYIERG KIDENV REIINHRSLRHPNI+
Sbjct: 241 DRYELVRDIGSGNFGVARLMKDKHTGELVAVKYIERGDKIDENVQREIINHRSLRHPNIV 420
Query: 62 RFKEVVLTPTHLGIVMEYAAGGELFDRIC 90
RFKEV+LTPTHL IVMEYA+GGELF+RIC
Sbjct: 421 RFKEVILTPTHLAIVMEYASGGELFERIC 507
>TC14602 UP|Q8GS56 (Q8GS56) Phosphoenolpyruvate carboxylase kinase, complete
Length = 1256
Score = 135 bits (340), Expect = 1e-32
Identities = 89/266 (33%), Positives = 132/266 (49%), Gaps = 7/266 (2%)
Frame = +2
Query: 3 KYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDENVAREIINHRSLR-----H 57
+Y++ ++IG G FG H + A+K I++ D + N H
Sbjct: 65 QYQLCEEIGRGRFGTIFRCFHPISTDTFAVKLIDKSLLADSTDRHCLENEPKYMSLLSPH 244
Query: 58 PNIIRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFS--EDEARYFFQQLISGVSYCH 115
PNI++ +V L +V+E L DRI +A S E EA +QL+ V++CH
Sbjct: 245 PNILQIFDVFEDDDVLSMVIELCQPLTLLDRIVAANGTSIPEVEAAGLMKQLLEAVAHCH 424
Query: 116 SMQICHRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRRE 175
+ + HRD+K +N L G LK+ DFG ++ R VGTP Y+APEVL RE
Sbjct: 425 RLGVAHRDVKPDNVLFGGGGD--LKLADFGSAEWFGDGRRMSGVVGTPYYVAPEVLMGRE 598
Query: 176 YDGKLADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRH 235
Y G+ DVWSCGV LY+ML G PF + F I + +I + ++S +
Sbjct: 599 Y-GEKVDVWSCGVILYIMLSGTPPFYGDSAAEIFEAVIRGNLRFPSRI--FRNVSPAAKD 769
Query: 236 LLSRIFVASPARRITIKEIKSHPWFL 261
LL ++ P+ RI+ ++ HPWFL
Sbjct: 770 LLRKMICRDPSNRISAEQALRHPWFL 847
>TC18664 similar to UP|Q944I6 (Q944I6) At2g30360/T9D9.17, partial (34%)
Length = 643
Score = 122 bits (305), Expect = 1e-28
Identities = 69/134 (51%), Positives = 85/134 (62%), Gaps = 4/134 (2%)
Frame = +3
Query: 3 KYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIER----GHKIDENVAREIINHRSLRHP 58
KYEV + +G G F R+ +T Q VA+K I + G + +V RE+ LRHP
Sbjct: 90 KYEVGRLLGCGAFAKVYHARNIETGQSVAVKVINKKKVIGTGLTGHVKREVSIMSRLRHP 269
Query: 59 NIIRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSMQ 118
NI+R EV+ T T + VME+A GGELF RI + GRFSED AR +FQQLIS V YCHS
Sbjct: 270 NIVRLHEVLATKTKIYFVMEFAKGGELFARISTKGRFSEDLARRYFQQLISAVGYCHSRG 449
Query: 119 ICHRDLKLENTLLD 132
+ HRDLK EN LLD
Sbjct: 450 VFHRDLKPENLLLD 491
>TC14018 similar to UP|Q43465 (Q43465) Protein kinase 2, partial (27%)
Length = 559
Score = 118 bits (295), Expect = 2e-27
Identities = 65/120 (54%), Positives = 85/120 (70%), Gaps = 5/120 (4%)
Frame = +3
Query: 210 RKTINRIMAVQYKIPDYVHISQECRHLLSRIFVASPARRITIKEIKSHPWFLKNLPRELT 269
RKTI RI+ VQY IPDYV +S ECR+LLSRIFVA PA+RITI EIK +PWFLK+LP+E+
Sbjct: 3 RKTIGRIIGVQYSIPDYVRVSAECRNLLSRIFVADPAKRITIAEIKQNPWFLKSLPKEII 182
Query: 270 EMAQAVYYRKENPTYSLQSIEDIMKIVEEAKNPPQ-ASRSVGG--FGWGG--EEDDDEIN 324
E + + S QS+E+IM+IV+EA+ P Q S+++ G G G EDD+EI+
Sbjct: 183 EAETKGFVETKKDQLS-QSVEEIMRIVQEARTPGQGGSKAIDGGQVGTGSMDVEDDEEID 359
>TC16440 similar to UP|Q93Y18 (Q93Y18) SNF1 related protein kinase, partial
(30%)
Length = 549
Score = 105 bits (261), Expect = 1e-23
Identities = 59/150 (39%), Positives = 86/150 (57%), Gaps = 5/150 (3%)
Frame = +1
Query: 3 KYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKIDEN----VAREIINHRSLRH- 57
KY++ + +G GNF VA+K I++ +D + + REI R L+H
Sbjct: 88 KYQLTRFLGRGNFAKVYQAVSLTDGTTVAVKMIDKSKTVDASMEPRIVREIDAMRRLQHH 267
Query: 58 PNIIRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHSM 117
PNI++ EV+ T T + ++++YA GGELF +I GR E AR +FQQL+S + +CH
Sbjct: 268 PNILKIHEVMATRTKIYLIVDYAGGGELFSKISRRGRLPEPLARRYFQQLVSALCFCHRN 447
Query: 118 QICHRDLKLENTLLDGSPAPRLKICDFGYS 147
+ HRDLK +N LLD LK+ DFG S
Sbjct: 448 GVAHRDLKPQNLLLDAE--GNLKVSDFGLS 531
>TC19452 homologue to UP|Q8S9J9 (Q8S9J9) At1g14000/F7A19_9, partial (41%)
Length = 562
Score = 94.4 bits (233), Expect = 3e-20
Identities = 62/153 (40%), Positives = 81/153 (52%), Gaps = 7/153 (4%)
Frame = +3
Query: 55 LRHPNIIRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYC 114
LRHPNI++F V L ++ EY GG+L + G S A F ++ G++Y
Sbjct: 69 LRHPNIVQFLGAVTERKPLMLITEYLRGGDLHQYLKEKGSLSPSTAINFSMDIVRGMAYL 248
Query: 115 HSMQ--ICHRDLKLENTLLDGSPAPRLKICDFGYSKSSLL---HSRPKST--VGTPAYIA 167
H+ I HRDLK N LL S A LK+ DFG SK + H K T G+ Y+A
Sbjct: 249 HNEPNVIIHRDLKPRNVLLVNSSADHLKVGDFGLSKLITVQNSHDVYKMTGETGSYRYMA 428
Query: 168 PEVLSRREYDGKLADVWSCGVTLYVMLVGAYPF 200
PEV R+YD K+ DV+S + LY ML G PF
Sbjct: 429 PEVFKHRKYDKKV-DVYSFAMILYEMLEGEPPF 524
>TC17375 mitogen-activated kinase kinase kinase alpha [Lotus corniculatus
var. japonicus]
Length = 1422
Score = 93.2 bits (230), Expect = 6e-20
Identities = 65/229 (28%), Positives = 110/229 (47%)
Frame = +3
Query: 57 HPNIIRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHS 116
HPNI+++ L L + +EY +GG + + G F E + + +Q++SG++Y HS
Sbjct: 108 HPNIVQYYGSELGEESLSVYLEYVSGGSIHKLLQEYGAFKEPVIQNYTRQIVSGLAYLHS 287
Query: 117 MQICHRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREY 176
HRD+K N L+D P +K+ DFG SK + S G+P ++APEV+
Sbjct: 288 RNTVHRDIKGANILVD--PNGEIKLADFGMSKHINSAASMLSFKGSPYWMAPEVVMNTNG 461
Query: 177 DGKLADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIPDYVHISQECRHL 236
G D+ S G T+ M P+ + K N +IP+ H+S + ++
Sbjct: 462 YGLPVDISSLGCTILEMATSKPPWSQFEGVAAIFKIGN--SKDMPEIPE--HLSDDAKNF 629
Query: 237 LSRIFVASPARRITIKEIKSHPWFLKNLPRELTEMAQAVYYRKENPTYS 285
+ + P R T + + +HP F+++ + T++A A R P S
Sbjct: 630 IKQCLQRDPLARPTAQSLLNHP-FIRD--QSATKVANASITRDAFPYMS 767
>CN825070
Length = 634
Score = 92.0 bits (227), Expect = 1e-19
Identities = 62/170 (36%), Positives = 81/170 (47%), Gaps = 6/170 (3%)
Frame = +3
Query: 2 EKYEVVKDIGSGNFGVARLMRHKDTKQLVAMKYIERGHKID----ENVAREI-INHRSLR 56
E Y + +G G FG L RH T A K I + + ++V REI I H
Sbjct: 123 EVYTFGRKLGQGQFGTTYLCRHNSTGCTFACKSIPKRKLLCKEDYDDVWREIQIMHHLSE 302
Query: 57 HPNIIRFKEVVLTPTHLGIVMEYAAGGELFDRICSAGRFSEDEARYFFQQLISGVSYCHS 116
HP+++R + IVME GGELFDRI G++SE EA + ++ V CHS
Sbjct: 303 HPHVVRIHGTYEDAASVHIVMEICEGGELFDRIVQKGQYSEREAAKLIRTIVEVVEACHS 482
Query: 117 MQICHRDLKLENTLLDG-SPAPRLKICDFGYSKSSLLHSRPKSTVGTPAY 165
+ + HRDLK EN L D +LK DFG S VG+P Y
Sbjct: 483 LGVMHRDLKPENFLFDTVEEDAKLKTTDFGLSVFYKPGETFGDVVGSPYY 632
>TC8079 homologue to UP|Q9M6S1 (Q9M6S1) MAP kinase 3, complete
Length = 1550
Score = 91.7 bits (226), Expect = 2e-19
Identities = 77/290 (26%), Positives = 124/290 (42%), Gaps = 34/290 (11%)
Frame = +3
Query: 7 VKDIGSGNFGVARLMRHKDTKQLVAMKYIERG---HKIDENVAREIINHRSLRHPNIIRF 63
+ IG G +G+ + + +T +LVA+K I H + REI R L H N+I
Sbjct: 300 IMPIGRGAYGIVCSLLNTETNELVAVKKIANAFDNHMDAKRTLREIKLLRHLDHENVIAL 479
Query: 64 KEVVLTPTHLGIVMEYAAGGELFDR-----ICSAGRFSEDEARYFFQQLISGVSYCHSMQ 118
++V+ P Y EL D I S SE+ +YF Q++ G+ Y HS
Sbjct: 480 RDVIPPPLRREFTDVYITT-ELMDTDLHQIIRSNQGLSEEHCQYFLYQVLRGLKYIHSAN 656
Query: 119 ICHRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPAYIAPEVLSRREYDG 178
I HRDLK N LL+ + LKI DFG ++ ++ + V T Y APE+L
Sbjct: 657 IIHRDLKPSNLLLNSN--CDLKIIDFGLARPTVENDFMTEYVVTRWYRAPELLLNSSDYT 830
Query: 179 KLADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQ-----------------Y 221
DVWS G +++ L+ P D + + + ++
Sbjct: 831 SAIDVWSVG-CIFMELMNKKPLFPGKDHVHQLRLLTELLGTPTEADLGLVKNDDVRRYIR 1007
Query: 222 KIPDY---------VHISQECRHLLSRIFVASPARRITIKEIKSHPWFLK 262
++P Y H+ L+ ++ P +RIT+++ +HP+ K
Sbjct: 1008QLPQYPRQPLTKVFPHVHPMAMDLVDKMLTIDPTKRITVEQALAHPYLEK 1157
>BP082166
Length = 378
Score = 82.4 bits (202), Expect = 1e-16
Identities = 38/57 (66%), Positives = 47/57 (81%)
Frame = -2
Query: 295 IVEEAKNPPQASRSVGGFGWGGEEDDDEINEAEAELEEDEYEKRVKEAQESGEIHVN 351
IVEEA+ P + RS+GG+GWG E+DD+ EAEAE+EEDEYEK VKEAQ SGEI+V+
Sbjct: 377 IVEEAQTPAKVPRSIGGYGWGEEDDDEAKEEAEAEVEEDEYEKSVKEAQASGEINVS 207
>TC9382 weakly similar to UP|ABL_DROME (P00522) Tyrosine-protein kinase Abl
(D-ash) , partial (5%)
Length = 1197
Score = 82.0 bits (201), Expect = 1e-16
Identities = 63/226 (27%), Positives = 104/226 (45%), Gaps = 1/226 (0%)
Frame = +2
Query: 46 AREIINHRSLRHPNIIRFKEVVLTPTHLGIVMEYAAGGELFDRI-CSAGRFSEDEARYFF 104
A+E+ R +RH N+++F +L IV E+ + G L+D + G F
Sbjct: 74 AQEVYIMRKIRHKNVVQFIGACTRTPNLCIVTEFMSRGSLYDFLHKQRGVFKLPSLLKVA 253
Query: 105 QQLISGVSYCHSMQICHRDLKLENTLLDGSPAPRLKICDFGYSKSSLLHSRPKSTVGTPA 164
+ G++Y H I HRDLK N L+D + +K+ DFG ++ + GT
Sbjct: 254 IDVSKGMNYLHQNNIIHRDLKTGNLLMDENEL--VKVADFGVARVITQSGVMTAETGTYR 427
Query: 165 YIAPEVLSRREYDGKLADVWSCGVTLYVMLVGAYPFEDQDDPKNFRKTINRIMAVQYKIP 224
++APEV+ + YD K ADV+S G+ L+ +L G P+ + + + ++ IP
Sbjct: 428 WMAPEVIEHKPYDQK-ADVFSFGIALWELLTGELPYSYLTPLQAAVGVVQK--GLRPSIP 598
Query: 225 DYVHISQECRHLLSRIFVASPARRITIKEIKSHPWFLKNLPRELTE 270
H LL R + P R EI L+N+ +E+ +
Sbjct: 599 KNTH--PRLSELLQRCWKQDPIERPAFSEIIE---ILQNIAKEVND 721
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.136 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,731,276
Number of Sequences: 28460
Number of extensions: 78084
Number of successful extensions: 726
Number of sequences better than 10.0: 180
Number of HSP's better than 10.0 without gapping: 641
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 660
length of query: 351
length of database: 4,897,600
effective HSP length: 91
effective length of query: 260
effective length of database: 2,307,740
effective search space: 600012400
effective search space used: 600012400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 56 (26.2 bits)
Medicago: description of AC134322.21


