

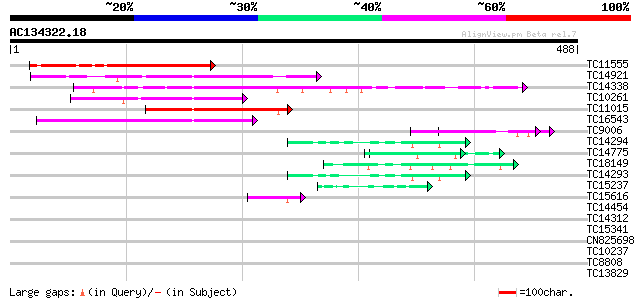
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC134322.18 - phase: 0
(488 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC11555 similar to UP|O49429 (O49429) DAG-like protein, partial ... 194 2e-50
TC14921 similar to UP|Q8L949 (Q8L949) Plastid protein, partial (... 124 3e-29
TC14338 similar to UP|Q9MAH5 (Q9MAH5) F12M16.16, partial (33%) 124 5e-29
TC10261 homologue to UP|DAG_ANTMA (Q38732) DAG protein, chloropl... 113 8e-26
TC11015 similar to PIR|T52623|T52623 DAG protein homolog [import... 112 2e-25
TC16543 110 4e-25
TC9006 81 3e-16
TC14294 similar to UP|Q9FYE4 (Q9FYE4) EF-hand calcium binding pr... 50 1e-06
TC14775 PIR|S14181|S14181 DNA-directed RNA polymerase largest c... 49 1e-06
TC18149 weakly similar to UP|Q91BL3 (Q91BL3) Essential structura... 47 7e-06
TC14293 similar to UP|Q9FYE4 (Q9FYE4) EF-hand calcium binding pr... 43 1e-04
TC15237 similar to UP|AAQ24632 (AAQ24632) GPRP, partial (64%) 42 3e-04
TC15616 similar to UP|DAG_ANTMA (Q38732) DAG protein, chloroplas... 41 5e-04
TC14454 similar to UP|O04133 (O04133) SRC2, partial (88%) 39 0.002
TC14312 similar to UP|Q8J174 (Q8J174) RfeF, partial (7%) 38 0.004
TC15341 similar to UP|Q9M0H8 (Q9M0H8) Predicted proline-rich pro... 37 0.006
CN825698 35 0.021
TC10237 similar to UP|NO75_SOYBN (P08297) Early nodulin 75 precu... 35 0.021
TC8808 35 0.036
TC13829 similar to UP|P93321 (P93321) Cdc2MsD protein, partial (... 34 0.061
>TC11555 similar to UP|O49429 (O49429) DAG-like protein, partial (14%)
Length = 467
Score = 194 bits (494), Expect = 2e-50
Identities = 109/160 (68%), Positives = 120/160 (74%)
Frame = +3
Query: 18 KTLLSPKPLLSTTTAMASSLILLRRSLRSLSGIHHSFSATATSLSVPITGSISSHFSLSK 77
KTLLS LS T +SSLI LRRSL +L+G H SFSAT SLS PI +SS +
Sbjct: 12 KTLLSLS--LSLTAMASSSLIRLRRSLTALAGFHRSFSAT--SLSAPIRYGVSS--PCWQ 173
Query: 78 SPNVVDGIQSRSFRSTSISLLSSRYGETSELSPEIGPDTILFEGCDYNHWLFVCDFPRDN 137
SPNV G QSRSF S+ SLLS R + + EIGPD ILFEGCDYNHWLFV DFPRDN
Sbjct: 174 SPNV--GFQSRSFMSSPTSLLSIRSSQRNIPDGEIGPDEILFEGCDYNHWLFVMDFPRDN 347
Query: 138 KPPPEEMIRIYEETCAKGLNISVEEAKKKIYACSTTTYTG 177
KPPPE+M+R YEETCAKGLNIS+EEAKKKIYACSTTTYTG
Sbjct: 348 KPPPEQMVRTYEETCAKGLNISLEEAKKKIYACSTTTYTG 467
>TC14921 similar to UP|Q8L949 (Q8L949) Plastid protein, partial (73%)
Length = 1030
Score = 124 bits (312), Expect = 3e-29
Identities = 93/254 (36%), Positives = 120/254 (46%), Gaps = 4/254 (1%)
Frame = +3
Query: 19 TLLSPKPLLSTTTAMASSLILLRRSLRSLSGIHHSFSATATSLSVPITGSISSHFSLSKS 78
TLL+ + +TTT +++ L + + S S S A + P T
Sbjct: 84 TLLTMRLFSTTTTTTTTAISLCKSPILSQSP-RRSVYALSQVFFTPTTR----------- 227
Query: 79 PNVVDGIQSRSFR---STSISLLSSRYGETSELSP-EIGPDTILFEGCDYNHWLFVCDFP 134
GI+ R R S L SS S+ P E+ P LF GCDYNHWL V D P
Sbjct: 228 ---FGGIRCRVNRAGDSAYSPLNSSGSNSFSDRPPTEMAP---LFPGCDYNHWLIVMDKP 389
Query: 135 RDNKPPPEEMIRIYEETCAKGLNISVEEAKKKIYACSTTTYTGFQAVMTEEESKKFEGIP 194
++MI Y +T AK L S EEAKKKIY S Y GF + EE S KFEG+
Sbjct: 390 GGEGATKQDMIDCYVQTLAKVLG-SEEEAKKKIYNVSCERYFGFGCEIDEETSNKFEGMN 566
Query: 195 GVIFVLPDSYIDPVNKQYGGDQYIEGQIIPRPPPVQFGRNLGGRRDYRQNNQLPNNRGNP 254
GV+FVLPDSY+DP K YG + ++ G+I+ RPP R R Q ++ P
Sbjct: 567 GVLFVLPDSYVDPEYKDYGAELFVNGEIVQRPP----------ERQRRVEPQQQRHQDRP 716
Query: 255 SYNNRDSMPRDGRN 268
YN+R R N
Sbjct: 717 RYNDRTRYVRRREN 758
>TC14338 similar to UP|Q9MAH5 (Q9MAH5) F12M16.16, partial (33%)
Length = 1751
Score = 124 bits (310), Expect = 5e-29
Identities = 127/422 (30%), Positives = 182/422 (43%), Gaps = 32/422 (7%)
Frame = +2
Query: 56 ATATSLSVPITGSIS--------SHFSLSKSPNVVDGIQSRSFRSTSISLLSSRYGETSE 107
+T+T+ + P ++S S S+ ++ +S S R+T+ SL + S
Sbjct: 101 STSTTTTTPSLSALSFLRRYRPLSAVSIPTCRHLFPSARSLSTRATTSSL-NDPNPNWSN 277
Query: 108 LSPEIGPDTILFEGCDYNHWLFVCDFPRDNKPPPEEMIRIYEETCAKGLNISVEEAKKKI 167
P+ +TIL +GCD+ HWL V + P + P +++I Y +T A+ + S +EA+ KI
Sbjct: 278 RPPK---ETILLDGCDFEHWLVVMEKP-EGDPTRDDIIDGYIKTLAQVVG-SEQEARMKI 442
Query: 168 YACSTTTYTGFQAVMTEEESKKFEGIPGVIFVLPDSYIDPVNKQYGGDQYIEGQIIPRPP 227
Y+ ST Y F A+++EE S K + +P V +VLPDSY++ K YGG+ +I G+ +P P
Sbjct: 443 YSVSTKHYFAFGALVSEELSYKLKELPKVRWVLPDSYLNVREKDYGGEPFINGEAVPYDP 622
Query: 228 PV--QFGRNLGGRRDYRQNNQLPNN--------RGNPSYNNRDSMPRDGRNY-GPPQN-- 274
++ RN + + N P N R + NRD R N GPP N
Sbjct: 623 KYHEEWVRNNARANERNRRNDRPRNNDRSRNFDRRRENVVNRDYQNRPPPNAGGPPSNQG 802
Query: 275 -FPPQQNHGQASHIP--PQQNIGQQQPNIR-------IDSTSQRFPPQQSYDQASQRYPP 324
FPP G S+ P N G + R S +PP +QA +P
Sbjct: 803 GFPPNNAGGPPSNQGGFPPNNAGGYPASPRPGNMPGPPPSNPGGYPPN---NQAGGYHPQ 973
Query: 325 QQSYDQASQGYLPKQNYGQAPQNYSAPQAPQNYSQQQTYGPASP-QYPPQQSFGPLGEGE 383
Q + GY P G P N A P N SQ Y P + YPP Q+
Sbjct: 974 NQ-----AGGYPPNSQAGGYPPNSQAGGYPPN-SQAGVYPPNNQGVYPPNQN-------- 1111
Query: 384 RRNYAPQQNFGPPGEVDRRNYAPQQNFGPPRQGERRNYVPQQNFGPPGQGERGNSVPSEG 443
NYA + G Y P N G P + NY PP Q G S
Sbjct: 1112--NYAGRTPPNVGGAPPNSAYGP--NAGYP--SNQNNYAGNMEGVPPNQNVGGYS--PNA 1267
Query: 444 GW 445
GW
Sbjct: 1268GW 1273
>TC10261 homologue to UP|DAG_ANTMA (Q38732) DAG protein, chloroplast
precursor, partial (41%)
Length = 510
Score = 113 bits (282), Expect = 8e-26
Identities = 66/160 (41%), Positives = 91/160 (56%), Gaps = 8/160 (5%)
Frame = +2
Query: 53 SFSATATSLSVPITGSISSHFSLSKSPNVVDGIQSRSFRSTSIS--------LLSSRYGE 104
+FS +A +L++P S F+ + + RS R+ + + L Y
Sbjct: 35 TFSFSAKTLTLPFHSKTPSLFNFFNPSLNFNRVAPRSIRAVTRARNPTRIRAALDEDYSA 214
Query: 105 TSELSPEIGPDTILFEGCDYNHWLFVCDFPRDNKPPPEEMIRIYEETCAKGLNISVEEAK 164
S E +TI+ GCDYNHWL V +FP+D P E+MI Y T + L S+EEAK
Sbjct: 215 KRSSSSE-QRETIMLPGCDYNHWLIVMEFPKDPAPSREQMIETYLFTLSTVLG-SMEEAK 388
Query: 165 KKIYACSTTTYTGFQAVMTEEESKKFEGIPGVIFVLPDSY 204
K +YA STTTYTGFQ + E S+KF+G+PGV++VLPDSY
Sbjct: 389 KNMYAFSTTTYTGFQCTVDEATSEKFKGLPGVLWVLPDSY 508
>TC11015 similar to PIR|T52623|T52623 DAG protein homolog [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(65%)
Length = 924
Score = 112 bits (279), Expect = 2e-25
Identities = 58/128 (45%), Positives = 78/128 (60%), Gaps = 2/128 (1%)
Frame = +1
Query: 118 LFEGCDYNHWLFVCDFPRDNKPPPEEMIRIYEETCAKGLNISVEEAKKKIYACSTTTYTG 177
LF GCDYNHWL + D P ++MI Y +T AK + S EEAKK+IY+ S Y G
Sbjct: 259 LFPGCDYNHWLIIMDKPGGEGATKKQMIDCYIKTLAKVVG-SEEEAKKRIYSISCEKYLG 435
Query: 178 FQAVMTEEESKKFEGIPGVIFVLPDSYIDPVNKQYGGDQYIEGQIIPRPPPVQ--FGRNL 235
F + EE S K +G G++FVLPDSY+DP + YGG+ ++ G I+ RPP Q FG+
Sbjct: 436 FGCEIDEETSDKLDGKAGIMFVLPDSYVDPEYQDYGGELFVNGAIVQRPPERQKRFGQKQ 615
Query: 236 GGRRDYRQ 243
R+ R+
Sbjct: 616 TTRKTSRK 639
>TC16543
Length = 575
Score = 110 bits (276), Expect = 4e-25
Identities = 71/191 (37%), Positives = 104/191 (54%), Gaps = 1/191 (0%)
Frame = +1
Query: 24 KPLLSTTTAMASSLILLRRSLRSL-SGIHHSFSATATSLSVPITGSISSHFSLSKSPNVV 82
+PL S T S + + R+L S + + S++ +S S P + ++ V
Sbjct: 4 EPLHSCTLHTL*SQMAYVNARRTLASTLSRALSSSPSSFSTPSRCRCIFALAAKQTLPVP 183
Query: 83 DGIQSRSFRSTSISLLSSRYGETSELSPEIGPDTILFEGCDYNHWLFVCDFPRDNKPPPE 142
D ++ +S S S + S +TIL +GCDY HWL V +FP + KP +
Sbjct: 184 DTVKFSVRTKSSGSGYSPLNDPSPNWSNRPPKETILLDGCDYEHWLIVMEFPENPKPSEQ 363
Query: 143 EMIRIYEETCAKGLNISVEEAKKKIYACSTTTYTGFQAVMTEEESKKFEGIPGVIFVLPD 202
EM+ Y +T + + S EEA KKIY+ ST TYTGF A+++EE S K + PGV++VLPD
Sbjct: 364 EMVNAYVKTLTQIVG-SEEEAMKKIYSVSTHTYTGFGALISEELSYKVKEXPGVLWVLPD 540
Query: 203 SYIDPVNKQYG 213
SY+D NK YG
Sbjct: 541 SYLDVPNKDYG 573
>TC9006
Length = 567
Score = 81.3 bits (199), Expect = 3e-16
Identities = 46/112 (41%), Positives = 58/112 (51%)
Frame = +3
Query: 346 QNYSAPQAPQNYSQQQTYGPASPQYPPQQSFGPLGEGERRNYAPQQNFGPPGEVDRRNYA 405
QNY + + + QQ Y P SPQ+ QQS GP G GERRN+ PQQ FG PG+ +RR+ A
Sbjct: 3 QNYPPQRGTYDSASQQRYAPPSPQFTQQQSAGPAGLGERRNFVPQQGFGQPGQGERRDLA 182
Query: 406 PQQNFGPPRQGERRNYVPQQNFGPPGQGERGNSVPSEGGWDFKPSYMEEFEQ 457
+ R PP + S + DFKPSYMEEFE+
Sbjct: 183 ARGVASEARGSTST---------PPYLKDFKPSYMEQSQEDFKPSYMEEFEK 311
Score = 65.9 bits (159), Expect = 1e-11
Identities = 44/113 (38%), Positives = 55/113 (47%), Gaps = 13/113 (11%)
Frame = +3
Query: 370 YPPQQSFGPLGEGERRNYAPQQNFGPPGEVDRRNYAPQQNFGPPRQGERRNYVPQQNFGP 429
YPPQ+ + A QQ + PP + QQ+ GP GERRN+VPQQ FG
Sbjct: 9 YPPQRG--------TYDSASQQRYAPPSP----QFTQQQSAGPAGLGERRNFVPQQGFGQ 152
Query: 430 PGQGER----GNSVPSEGGW---------DFKPSYMEEFEQGQKGNNHAEEQK 469
PGQGER V SE DFKPSYME+ ++ K + E +K
Sbjct: 153 PGQGERRDLAARGVASEARGSTSTPPYLKDFKPSYMEQSQEDFKPSYMEEFEK 311
Score = 33.5 bits (75), Expect = 0.080
Identities = 25/77 (32%), Positives = 37/77 (47%), Gaps = 13/77 (16%)
Frame = +3
Query: 307 QRFPPQQ-SYDQASQ-RYPP-------QQSYDQASQG----YLPKQNYGQAPQNYSAPQA 353
Q +PPQ+ +YD ASQ RY P QQS A G ++P+Q +GQ Q A
Sbjct: 3 QNYPPQRGTYDSASQQRYAPPSPQFTQQQSAGPAGLGERRNFVPQQGFGQPGQGERRDLA 182
Query: 354 PQNYSQQQTYGPASPQY 370
+ + + ++P Y
Sbjct: 183 ARGVASEARGSTSTPPY 233
>TC14294 similar to UP|Q9FYE4 (Q9FYE4) EF-hand calcium binding protein-like
(At5g04170), partial (65%)
Length = 1192
Score = 49.7 bits (117), Expect = 1e-06
Identities = 51/166 (30%), Positives = 63/166 (37%), Gaps = 9/166 (5%)
Frame = +1
Query: 240 DYRQNNQLPNNRGNPSYNNRDSMPRDGRNYGPPQNFPPQQNHGQASHIPPQQNIGQQQPN 299
DY ++P++ Y N+ S G YG P PP Q +G A PP Q G
Sbjct: 22 DYSSFFKIPSHFNMSGYPNKPS----GYGYGAP---PPYQPYGAA---PPSQPYGAA--- 162
Query: 300 IRIDSTSQRFPPQQSYDQASQRYPPQQSYDQASQGYLPKQNYGQAP--QNYSAPQAPQNY 357
PP QSY PP Q Y A P Q YG P Q+Y P P
Sbjct: 163 ----------PPSQSYGAP----PPPQPYGAAP----PSQPYGAPPPSQSYGGPPPPSQP 288
Query: 358 SQQQTYG-PASP-----QYPPQQSFGPLGEGERRNYAPQQN-FGPP 396
YG P++P Q PP+ G G Y P + +G P
Sbjct: 289 YSASPYGQPSAPYAAPYQKPPKDESHSHGGGGGSGYPPPPSAYGSP 426
>TC14775 PIR|S14181|S14181 DNA-directed RNA polymerase largest chain
(isoform B1) - soybean (fragment) {Glycine
max;} , partial (22%)
Length = 848
Score = 49.3 bits (116), Expect = 1e-06
Identities = 35/118 (29%), Positives = 45/118 (37%), Gaps = 1/118 (0%)
Frame = +1
Query: 310 PPQQSYDQASQRYPPQQSYDQASQGYLPKQNYGQAPQNYSAPQAPQNYSQQQTYGPASPQ 369
P SY+ S +Y P +Y +S P Y NYS P +P +Y P+SP
Sbjct: 97 PTSPSYNPQSAKYSPSLAYSPSSPRLSPSSPYSPTSPNYS-PTSPSYSPTSPSYSPSSPT 273
Query: 370 YPPQQSFGPLGEGERRNYAPQQ-NFGPPGEVDRRNYAPQQNFGPPRQGERRNYVPQQN 426
Y P P G +Y+P F P Y+P Q P Y PQ N
Sbjct: 274 YSPS---SPYNSGVSPDYSPSSPQFSP-----STGYSPSQPGYSP--SSTSQYTPQTN 417
Score = 47.8 bits (112), Expect = 4e-06
Identities = 28/98 (28%), Positives = 39/98 (39%), Gaps = 11/98 (11%)
Frame = +1
Query: 306 SQRFPPQQSYDQASQRYPPQQSYDQASQGYLPKQ-NYGQAPQNYS------APQAPQNYS 358
S ++ P +Y +S R P Y S Y P +Y +YS +P +P N
Sbjct: 124 SAKYSPSLAYSPSSPRLSPSSPYSPTSPNYSPTSPSYSPTSPSYSPSSPTYSPSSPYNSG 303
Query: 359 QQQTYGPASPQYPPQQSFGPLGEG----ERRNYAPQQN 392
Y P+SPQ+ P + P G Y PQ N
Sbjct: 304 VSPDYSPSSPQFSPSTGYSPSQPGYSPSSTSQYTPQTN 417
Score = 27.3 bits (59), Expect = 5.7
Identities = 33/129 (25%), Positives = 42/129 (31%), Gaps = 5/129 (3%)
Frame = +1
Query: 327 SYDQASQGYLPKQNYGQAPQNYSAPQAPQNYSQQQTYGPASPQYPPQQSFGPLGEGERRN 386
SY S Y P YS P +P +Y P SP Y PQ +
Sbjct: 4 SYSPTSPAYSPTS------PGYS-PTSPSYSPTSPSYSPTSPSYNPQSA----------K 132
Query: 387 YAPQQNFGP--PGEVDRRNYAP-QQNFGPPRQGERRNYVPQQNFGPPGQGERGNSVPSEG 443
Y+P + P P Y+P N+ P +Y P P S P
Sbjct: 133 YSPSLAYSPSSPRLSPSSPYSPTSPNYSPTSP----SYSPTSPSYSPSSPTYSPSSPYNS 300
Query: 444 GW--DFKPS 450
G D+ PS
Sbjct: 301 GVSPDYSPS 327
>TC18149 weakly similar to UP|Q91BL3 (Q91BL3) Essential structural protein
pp78/81, partial (5%)
Length = 710
Score = 47.0 bits (110), Expect = 7e-06
Identities = 58/187 (31%), Positives = 71/187 (37%), Gaps = 19/187 (10%)
Frame = +3
Query: 271 PPQNFPPQQNHGQASHIPPQQNIGQQQPNIRIDSTSQ--RFPPQQSYDQASQRYPPQQSY 328
PP N PPQ +PPQQ + P S +Q P + Y PP QS
Sbjct: 117 PPPNAPPQ--------LPPQQGLPPSFPPPNQFSQTQIPAVPQRDPYFP-----PPVQSQ 257
Query: 329 DQASQGYLPKQNYGQ--APQNYSAPQAPQNYSQQQT-----YGPASPQY-PPQQSFGP-- 378
+ P Q Y Q APQ + P AP + QQT PA PQ+ PP S P
Sbjct: 258 ET------PNQQYQQPLAPQPHPQPGAPPHQQYQQTPHPQYSQPAPPQHQPPIPSINPPQ 419
Query: 379 ----LGEG-ERRNYAPQQNFGPPGEVDRRNYAPQQNFGPPRQGERRNY--VPQQNFGPPG 431
LG E Y P Q + P + R Q GPP ++ Y P PP
Sbjct: 420 LQSSLGHHVEEPPYVPSQTYPP----NLRQPPSQPPSGPPAPPSQQFYGTAPHAYEPPPS 587
Query: 432 QGERGNS 438
+ G S
Sbjct: 588 RSGSGYS 608
>TC14293 similar to UP|Q9FYE4 (Q9FYE4) EF-hand calcium binding protein-like
(At5g04170), partial (18%)
Length = 540
Score = 42.7 bits (99), Expect = 1e-04
Identities = 47/166 (28%), Positives = 59/166 (35%), Gaps = 9/166 (5%)
Frame = +2
Query: 240 DYRQNNQLPNNRGNPSYNNRDSMPRDGRNYGPPQNFPPQQNHGQASHIPPQQNIGQQQPN 299
DY ++P++ Y N+ S G YG P PP Q +G A
Sbjct: 62 DYSSFFKIPSHFNMSGYPNKPS----GYGYGAP---PPYQPYGAA--------------- 175
Query: 300 IRIDSTSQRFPPQQSYDQASQRYPPQQSYDQASQGYLPKQNYGQAP--QNYSAPQAPQNY 357
PP QSY PP Q Y A P Q YG P Q+Y P P
Sbjct: 176 ----------PPSQSYGAP----PPPQPYGAAP----PSQPYGAPPPSQSYGGPPPPSQP 301
Query: 358 SQQQTYG-PASP-----QYPPQQSFGPLGEGERRNYAPQQN-FGPP 396
YG P++P Q PP+ G G Y P + +G P
Sbjct: 302 YSASPYGQPSAPYAAPYQKPPKDESHSHGGGGGSGYPPPPSAYGSP 439
>TC15237 similar to UP|AAQ24632 (AAQ24632) GPRP, partial (64%)
Length = 814
Score = 41.6 bits (96), Expect = 3e-04
Identities = 34/99 (34%), Positives = 39/99 (39%)
Frame = +2
Query: 266 GRNYGPPQNFPPQQNHGQASHIPPQQNIGQQQPNIRIDSTSQRFPPQQSYDQASQRYPPQ 325
G Y PP +PPQ HG PPQQ +PPQQ YPPQ
Sbjct: 197 GHGY-PPGAYPPQ--HG----YPPQQG----------------YPPQQG-------YPPQ 286
Query: 326 QSYDQASQGYLPKQNYGQAPQNYSAPQAPQNYSQQQTYG 364
Q Y A GY P+Q Y P Y P + Q +G
Sbjct: 287 QGYPPA--GYPPQQGY--PPAGYPGPSGAHGHGGHQGHG 391
Score = 34.3 bits (77), Expect = 0.047
Identities = 18/48 (37%), Positives = 23/48 (47%)
Frame = +2
Query: 365 PASPQYPPQQSFGPLGEGERRNYAPQQNFGPPGEVDRRNYAPQQNFGP 412
P YPPQQ + P ++ Y PQQ + P G Y PQQ + P
Sbjct: 224 PPQHGYPPQQGYPP-----QQGYPPQQGYPPAG------YPPQQGYPP 334
Score = 34.3 bits (77), Expect = 0.047
Identities = 23/80 (28%), Positives = 32/80 (39%), Gaps = 7/80 (8%)
Frame = +2
Query: 325 QQSYDQASQGYLPKQNYGQAPQNYSAPQAP-------QNYSQQQTYGPASPQYPPQQSFG 377
+ +D++ +G +G A + P Y QQ Y P YPPQQ +
Sbjct: 122 KDKHDESDKGVFSSLAHGVANAAHGGHGYPPGAYPPQHGYPPQQGY-PPQQGYPPQQGYP 298
Query: 378 PLGEGERRNYAPQQNFGPPG 397
P G Y PQQ + P G
Sbjct: 299 PAG------YPPQQGYPPAG 340
Score = 32.0 bits (71), Expect = 0.23
Identities = 18/46 (39%), Positives = 24/46 (52%), Gaps = 1/46 (2%)
Frame = +2
Query: 387 YAPQQNFGPPGEVDRRNYAPQQNFGPPRQG-ERRNYVPQQNFGPPG 431
Y PQ + P ++ Y PQQ + PP+QG Y PQQ + P G
Sbjct: 221 YPPQHGYPP-----QQGYPPQQGY-PPQQGYPPAGYPPQQGYPPAG 340
Score = 30.0 bits (66), Expect = 0.89
Identities = 17/38 (44%), Positives = 21/38 (54%), Gaps = 1/38 (2%)
Frame = +2
Query: 395 PPGEVD-RRNYAPQQNFGPPRQGERRNYVPQQNFGPPG 431
PPG + Y PQQ + PP+QG Y PQQ + P G
Sbjct: 209 PPGAYPPQHGYPPQQGY-PPQQG----YPPQQGYPPAG 307
>TC15616 similar to UP|DAG_ANTMA (Q38732) DAG protein, chloroplast
precursor, partial (13%)
Length = 650
Score = 40.8 bits (94), Expect = 5e-04
Identities = 23/55 (41%), Positives = 29/55 (51%), Gaps = 5/55 (9%)
Frame = +3
Query: 205 IDPVNKQYGGDQYIEGQIIPRPPPVQFGRNLGG-----RRDYRQNNQLPNNRGNP 254
ID NK YGGD+YI G+IIP P + GG RR R+ + P +R P
Sbjct: 3 IDVKNKDYGGDKYINGEIIPSKYPTYQPKRSGGSRNDSRRYERKRDGPPTDRRRP 167
>TC14454 similar to UP|O04133 (O04133) SRC2, partial (88%)
Length = 1190
Score = 38.9 bits (89), Expect = 0.002
Identities = 31/97 (31%), Positives = 38/97 (38%), Gaps = 2/97 (2%)
Frame = +1
Query: 342 GQAPQNYSAPQAPQNYSQQQTYGPASPQYPPQQSFGPLGEGERRNYAPQQ-NFGPPGEVD 400
G + Y PQAP YG P Y Q G + Y PQQ +G P +
Sbjct: 679 GSSSVPYGTPQAPG-------YGYPPPAYGAQPGHGYPPQQPGYGYPPQQAGYGYPPQQP 837
Query: 401 RRNYAPQQ-NFGPPRQGERRNYVPQQNFGPPGQGERG 436
Y PQQ +G P Q Y PQQ P + + G
Sbjct: 838 GYGYPPQQAGYGYPPQQAGYGYPPQQAGQKPKKNQLG 948
Score = 37.4 bits (85), Expect = 0.006
Identities = 30/74 (40%), Positives = 33/74 (44%), Gaps = 7/74 (9%)
Frame = +1
Query: 309 FPPQQSYDQASQRYPPQQSY-----DQASQGYLPKQ-NYGQAPQNYSAPQAPQNYSQQQT 362
+PP Q YPPQQ QA GY P+Q YG PQ QA Y QQ
Sbjct: 727 YPPPAYGAQPGHGYPPQQPGYGYPPQQAGYGYPPQQPGYGYPPQ-----QAGYGYPPQQA 891
Query: 363 -YGPASPQYPPQQS 375
YG YPPQQ+
Sbjct: 892 GYG-----YPPQQA 918
>TC14312 similar to UP|Q8J174 (Q8J174) RfeF, partial (7%)
Length = 622
Score = 37.7 bits (86), Expect = 0.004
Identities = 32/123 (26%), Positives = 48/123 (39%), Gaps = 1/123 (0%)
Frame = +1
Query: 269 YGPPQNFPPQQNHGQASHIPPQQN-IGQQQPNIRIDSTSQRFPPQQSYDQASQRYPPQQS 327
YG Q+ +GQ+++ P Q + G QP+ + P Y A+Q QS
Sbjct: 43 YGQSQSPSTAGGYGQSAYPPAQPSPAGYAQPDQKA--------PASGYGVAAQPGYGPQS 198
Query: 328 YDQASQGYLPKQNYGQAPQNYSAPQAPQNYSQQQTYGPASPQYPPQQSFGPLGEGERRNY 387
Y P+ +YGQAP +Y Y+Q Y P Q+ +G
Sbjct: 199 YGA------PQGSYGQAPPSYGNSSYGAGYAQTPAYASDGTNGAPAQAAAAAQQGGVAKA 360
Query: 388 APQ 390
+PQ
Sbjct: 361 SPQ 369
>TC15341 similar to UP|Q9M0H8 (Q9M0H8) Predicted proline-rich protein,
partial (33%)
Length = 1002
Score = 37.4 bits (85), Expect = 0.006
Identities = 37/117 (31%), Positives = 42/117 (35%), Gaps = 14/117 (11%)
Frame = +1
Query: 272 PQNFPPQQNHGQASHIPPQQNIGQ--QQPNIRIDST-------SQRFPPQQSYDQASQRY 322
P PQ A P N+ Q QQP I ST + PQ Y Q+Y
Sbjct: 574 PNQIAPQPQPAAAQPQAPAPNVTQAPQQPAYYIPSTPLPGNTQTVAQLPQNQYLPPDQQY 753
Query: 323 PPQQSYDQASQGYLPKQNY-GQAP----QNYSAPQAPQNYSQQQTYGPASPQYPPQQ 374
Q D + P N G +P Q Y Q PQ QQQ PQ PQQ
Sbjct: 754 RTPQMQDMSRVAPQPCTNXSGXSPPPPVQQYPQYQQPQQQQQQQQQQQQWPQQLPQQ 924
>CN825698
Length = 704
Score = 35.4 bits (80), Expect = 0.021
Identities = 30/112 (26%), Positives = 48/112 (42%), Gaps = 6/112 (5%)
Frame = +1
Query: 330 QASQGYLPKQNYGQAPQNY---SAPQAPQNYSQQQTYGPASPQY---PPQQSFGPLGEGE 383
+ SQ +P AP + S P +P + S P P PP S P
Sbjct: 34 RVSQFPIPMAEVINAPPDSLDKSPPPSPSSASAPPPPPPPPPPPSLPPPSDSDLPPPPAR 213
Query: 384 RRNYAPQQNFGPPGEVDRRNYAPQQNFGPPRQGERRNYVPQQNFGPPGQGER 435
RR+ ++F P +RR+Y + N PP + R++ +++ PP Q +R
Sbjct: 214 RRDRRDDRDFDRPP--NRRDYYDRSNNSPPPKDRDRDFKRRRSPSPPSQRDR 363
Score = 27.7 bits (60), Expect = 4.4
Identities = 21/66 (31%), Positives = 30/66 (44%), Gaps = 2/66 (3%)
Frame = +1
Query: 226 PPPVQFGRNLGGRRDYRQNNQLPNNRGNPSYNNRDSMPRD-GRNYGPPQN-FPPQQNHGQ 283
PPP R+ RRD R ++ PN R +N P+D R++ ++ PP Q
Sbjct: 196 PPPPARRRD---RRDDRDFDRPPNRRDYYDRSNNSPPPKDRDRDFKRRRSPSPPSQRDRD 366
Query: 284 ASHIPP 289
H PP
Sbjct: 367 RRHSPP 384
Score = 26.9 bits (58), Expect = 7.5
Identities = 15/39 (38%), Positives = 17/39 (43%)
Frame = +3
Query: 252 GNPSYNNRDSMPRDGRNYGPPQNFPPQQNHGQASHIPPQ 290
G P + + PR R P Q PP Q GQ PPQ
Sbjct: 228 GRPGFRSST*PPRLLR---PQQQLPPAQRPGQGLQAPPQ 335
>TC10237 similar to UP|NO75_SOYBN (P08297) Early nodulin 75 precursor (N-75)
(NGM-75), partial (49%)
Length = 803
Score = 35.4 bits (80), Expect = 0.021
Identities = 38/162 (23%), Positives = 57/162 (34%), Gaps = 4/162 (2%)
Frame = +1
Query: 269 YGPPQNFPPQQNHGQASHIPPQQNIGQQQPNIRIDSTSQRFPPQQSYDQASQRYPPQQSY 328
Y PP PP Q + PPQ ++ P + + + P +Y ++ PP+
Sbjct: 94 YQPPHKKPPSQYQPPPEYEPPQ----EKPPPVYL---PPYWKPPPAYPPPYEKPPPEYEP 252
Query: 329 DQASQGYLPKQNYGQAPQNYSAPQAPQNYSQQQTYGP----ASPQYPPQQSFGPLGEGER 384
Q + Y + P + P P Y Y P P Y P P+ E
Sbjct: 253 PQEKPPPVYSPPYERPPTGHPTPY-PPFYHLPPVYHPPYEKPPPLYQPPHEKPPIYEPP- 426
Query: 385 RNYAPQQNFGPPGEVDRRNYAPQQNFGPPRQGERRNYVPQQN 426
+ + PP E Y P + PP G +Y P +N
Sbjct: 427 --HEKPPIYEPPHEKPPL-YEPPPGYNPPPYG---HYPPSKN 534
>TC8808
Length = 579
Score = 34.7 bits (78), Expect = 0.036
Identities = 38/132 (28%), Positives = 49/132 (36%), Gaps = 2/132 (1%)
Frame = +1
Query: 285 SHIPPQQNIGQQQPNIRIDSTSQRFPPQQSYDQASQRYPPQQSYDQASQGYLPKQNYGQA 344
+H P QP + + P SY Q YP QQ+Y + S Y QA
Sbjct: 7 NHPPAASGPVYAQPGGQPTYSQPSAQPAASYAQGYGSYPSQQTYPEQSAPNNAVYGY-QA 183
Query: 345 PQ--NYSAPQAPQNYSQQQTYGPASPQYPPQQSFGPLGEGERRNYAPQQNFGPPGEVDRR 402
PQ +Y + AP YS Q+ P Q P Q+ G + YA P
Sbjct: 184 PQDPSYGSAAAPA-YSAAQSGQPGYAQPTPTQT----GYEQSAGYAAVPAASAPAAA-YP 345
Query: 403 NYAPQQNFGPPR 414
Y Q +G PR
Sbjct: 346 QYDTAQVYGAPR 381
>TC13829 similar to UP|P93321 (P93321) Cdc2MsD protein, partial (43%)
Length = 515
Score = 33.9 bits (76), Expect = 0.061
Identities = 35/124 (28%), Positives = 50/124 (40%), Gaps = 20/124 (16%)
Frame = +3
Query: 236 GGRRDYRQNNQLPNNRGNPSYNNRDSMPRDGRNYGP----PQNFPPQQNHGQASHIPP-- 289
G RR RQ+ Q PS ++++ RDGR P P++ PP H PP
Sbjct: 108 GRRRHIRQSLQSQREIHRPSRRSQENPSRDGRRRRPSDRPPRSLPPPDALPIHLHRPPPQ 287
Query: 290 ----QQNIGQQQP----NIRIDSTSQRFPP------QQSYDQASQRYPPQQSYDQASQGY 335
+QN +QQP R S++ PP + Q RY PQ + A+ Y
Sbjct: 288 SRARRQNPQKQQPPPLRRRRPPSSAV*TPPLPRLRVPRHRSQEIHRYLPQGAKP*AASAY 467
Query: 336 LPKQ 339
+
Sbjct: 468 FDSE 479
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.312 0.133 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,384,305
Number of Sequences: 28460
Number of extensions: 161494
Number of successful extensions: 930
Number of sequences better than 10.0: 105
Number of HSP's better than 10.0 without gapping: 834
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 887
length of query: 488
length of database: 4,897,600
effective HSP length: 94
effective length of query: 394
effective length of database: 2,222,360
effective search space: 875609840
effective search space used: 875609840
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 57 (26.6 bits)
Medicago: description of AC134322.18


