

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC134049.16 - phase: 0
(166 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
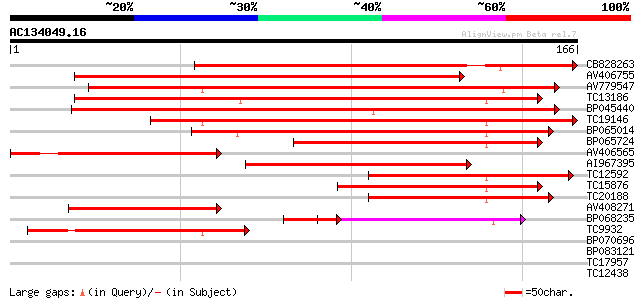
Score E
Sequences producing significant alignments: (bits) Value
CB828263 158 5e-40
AV406755 138 4e-34
AV779547 135 3e-33
TC13186 weakly similar to UP|Q8W2C0 (Q8W2C0) Functional candidat... 117 8e-28
BP045440 110 9e-26
TC19146 similar to UP|Q8S4X3 (Q8S4X3) TIR-similar-domain-contain... 102 3e-23
BP065014 91 1e-19
BP065724 77 2e-15
AV406565 74 2e-14
AI967395 72 6e-14
TC12592 54 1e-08
TC15876 weakly similar to UP|Q8S4X3 (Q8S4X3) TIR-similar-domain-... 53 2e-08
TC20188 weakly similar to UP|Q8S4X3 (Q8S4X3) TIR-similar-domain-... 50 2e-07
AV408271 50 2e-07
BP068235 45 3e-07
TC9932 weakly similar to UP|Q93YA7 (Q93YA7) Resistance gene-like... 46 3e-06
BP070696 33 0.019
BP083121 28 1.1
TC17957 weakly similar to GB|AAM98310.1|22655436|AY142046 At2g19... 27 1.4
TC12438 similar to UP|ALG6_ARATH (Q9FF17) Probable dolichyl pyro... 27 1.8
>CB828263
Length = 570
Score = 158 bits (399), Expect = 5e-40
Identities = 78/113 (69%), Positives = 93/113 (82%), Gaps = 1/113 (0%)
Frame = +2
Query: 55 DLNSGDEIPTTLVRAIEEAKLSVIVFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYD 114
DL GDEI ++L+RAIEEAKLSVIVFSKNY SKWCL+EL+KILE K+ +GQIV+PVFYD
Sbjct: 5 DLQRGDEISSSLLRAIEEAKLSVIVFSKNYGNSKWCLDELVKILECKRTRGQIVLPVFYD 184
Query: 115 VDPSDVRNQRGSYAEAFAKHENNFEGKI-KVQEWRNGLLEAANYAGWDCNVNR 166
+DPS VRNQ G+YAEAF KH G++ KVQ+WR L EAAN +GWDC+VNR
Sbjct: 185 IDPSHVRNQTGTYAEAFVKH-----GQVDKVQKWREALREAANLSGWDCSVNR 328
>AV406755
Length = 425
Score = 138 bits (348), Expect = 4e-34
Identities = 61/114 (53%), Positives = 92/114 (80%)
Frame = +3
Query: 20 HEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDNDLNSGDEIPTTLVRAIEEAKLSVIV 79
++VF+SFRGE++R +FTSHL ALK I+ ++DN+L G++I ++L++AIE++++++I+
Sbjct: 84 YDVFLSFRGEESRRSFTSHLYTALKNAGIKVFMDNELQRGEDISSSLLKAIEDSRIAIII 263
Query: 80 FSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSDVRNQRGSYAEAFAK 133
FS NY SKWCL+EL KI+E ++ GQ V+PVFY+VDPSD+R QRG+ EAF K
Sbjct: 264 FSTNYTGSKWCLDELEKIIECQRTIGQEVMPVFYNVDPSDIRKQRGTVGEAFRK 425
>AV779547
Length = 556
Score = 135 bits (341), Expect = 3e-33
Identities = 72/140 (51%), Positives = 92/140 (65%), Gaps = 2/140 (1%)
Frame = +1
Query: 24 ISFRGEDTRNNFTSHLNGALKRLDIRTYIDND-LNSGDEIPTTLVRAIEEAKLSVIVFSK 82
+SFRGEDTRNNFT HL GAL T+ D+ L G I T L++AIE +++ ++VFSK
Sbjct: 109 VSFRGEDTRNNFTDHLLGALYGKGFVTFKDDTMLRKGKNISTELIQAIEGSQILIVVFSK 288
Query: 83 NYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSDVRNQRGSYAEAFAKHENNFEGKI 142
YA S WCL+EL KI + K Q V+PV DV PS+VR Q G+Y EAF KHE F+ +
Sbjct: 289 YYASSTWCLQELAKIADGIVGKRQTVLPVVCDVTPSEVRKQSGNYGEAFLKHEERFKEDL 468
Query: 143 K-VQEWRNGLLEAANYAGWD 161
+ VQ WR L E A+ +GWD
Sbjct: 469 EMVQRWRKALAEVADLSGWD 528
>TC13186 weakly similar to UP|Q8W2C0 (Q8W2C0) Functional candidate
resistance protein KR1, partial (6%)
Length = 586
Score = 117 bits (294), Expect = 8e-28
Identities = 62/139 (44%), Positives = 93/139 (66%), Gaps = 2/139 (1%)
Frame = +1
Query: 20 HEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDNDLNSGDEIPTTL-VRAIEEAKLSVI 78
++VFIS DTR F+ L AL I T+ID+ L D+I +T+ +AIE ++++++
Sbjct: 103 YDVFISLITSDTRFGFSGDLYTALSDKGILTFIDDGLPRPDDIMSTVNSKAIESSRIAIV 282
Query: 79 VFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSDVRNQRGSYAEAFAKHENNF 138
V S+NYA S +CL+ELM IL+ ++ KG ++ PVFY V+PSDVR Q+G + +AFA HE F
Sbjct: 283 VISENYASSSFCLDELMYILKRREEKGLLIWPVFYQVEPSDVRYQKGGFGKAFASHEERF 462
Query: 139 -EGKIKVQEWRNGLLEAAN 156
KV++WRN L + A+
Sbjct: 463 GSDSAKVRKWRNALKQVAD 519
>BP045440
Length = 535
Score = 110 bits (276), Expect = 9e-26
Identities = 58/145 (40%), Positives = 88/145 (60%), Gaps = 2/145 (1%)
Frame = -3
Query: 19 KHEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDNDLNSGDEIPTTLVRAIEEAKLSVI 78
++++FISFRG DTR++FT L L R T+ D++ G L+ IEE++ ++I
Sbjct: 530 RYQIFISFRGMDTRDDFTKPLYQGLCREGFVTFKDDESLEGGSPIEKLLDDIEESRFAII 351
Query: 79 VFSKNYAVSKWCLEELMKILEIKKMKG--QIVVPVFYDVDPSDVRNQRGSYAEAFAKHEN 136
+ SKNYA S+WCL+EL KILE K +G Q+V+P +V P++VR+ YA+A AKHE
Sbjct: 350 ILSKNYAESEWCLKELSKILECKDQEGKNQLVLPSSTEVTPTEVRHVINRYADAMAKHEK 171
Query: 137 NFEGKIKVQEWRNGLLEAANYAGWD 161
N +Q W+ L + G++
Sbjct: 170 NGIDSKTIQAWKKALFDVCTLTGFE 96
>TC19146 similar to UP|Q8S4X3 (Q8S4X3) TIR-similar-domain-containing protein
TSDC, partial (12%)
Length = 701
Score = 102 bits (255), Expect = 3e-23
Identities = 53/127 (41%), Positives = 83/127 (64%), Gaps = 2/127 (1%)
Frame = +1
Query: 42 ALKRLDIRTYIDND-LNSGDEIPTTLVRAIEEAKLSVIVFSKNYAVSKWCLEELMKILEI 100
AL+R +T++D++ L G+EI +L+ A E+++LS+IVFS+NY S WCL+EL+KI+E
Sbjct: 10 ALRREGFKTFMDDEGLVGGNEIAKSLLDASEKSRLSIIVFSENYGYSSWCLDELVKIVEC 189
Query: 101 KKMKGQIVVPVFYDVDPSDVRNQRGSYAEAFAKHENNF-EGKIKVQEWRNGLLEAANYAG 159
+ K Q+V P+FY V+ SDV +Q SY A HE N+ + +V++WR+ L A G
Sbjct: 190 METKKQLVWPIFYKVEKSDVTSQENSYEHAMIGHEKNYGKDSDEVKKWRSALSAMAKLEG 369
Query: 160 WDCNVNR 166
+ N+
Sbjct: 370 HNVKENK 390
>BP065014
Length = 498
Score = 90.5 bits (223), Expect = 1e-19
Identities = 44/108 (40%), Positives = 73/108 (66%), Gaps = 2/108 (1%)
Frame = -2
Query: 54 NDLNSGDEIPTT-LVRAIEEAKLSVIVFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVF 112
+DL+ D P L+ AIEE++++++V S+N+A SKWCL+EL+KIL+ +K K +V+P+F
Sbjct: 491 DDLSLEDGAPIEELLGAIEESRVAIVVLSENFADSKWCLKELVKILDCRKKKKPLVLPIF 312
Query: 113 YDVDPSDVRNQRGSYAEAFAKHENNF-EGKIKVQEWRNGLLEAANYAG 159
Y V+P+++R+ +G Y EA A+HE + V+EW L + + G
Sbjct: 311 YKVEPTNIRHLKGRYGEAMAEHEKKLGKDSQTVREWTQALSDISYLKG 168
>BP065724
Length = 495
Score = 76.6 bits (187), Expect = 2e-15
Identities = 36/74 (48%), Positives = 51/74 (68%), Gaps = 1/74 (1%)
Frame = -1
Query: 84 YAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSDVRNQRGSYAEAFAKHENNF-EGKI 142
YA S WCLEE++KILE K+ Q+V P+FY V+PSDVR+QR SY +A K E + +
Sbjct: 495 YADSSWCLEEVVKILECKQKNDQLVWPIFYKVEPSDVRHQRNSYEKAMIKQETRYGKDSE 316
Query: 143 KVQEWRNGLLEAAN 156
KV++WR+ L + +
Sbjct: 315 KVKKWRSSLSQVCD 274
>AV406565
Length = 207
Score = 73.6 bits (179), Expect = 2e-14
Identities = 37/62 (59%), Positives = 43/62 (68%)
Frame = +2
Query: 1 MAWSYSSSSSSFNNAPLEKHEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDNDLNSGD 60
MAWS +SSS AP +KH+VF+SFRGEDTR T HL L RL ++TYID DL GD
Sbjct: 35 MAWSTTSSS-----APQQKHDVFLSFRGEDTRYTCTGHLQATLTRLQVKTYIDYDLQRGD 199
Query: 61 EI 62
EI
Sbjct: 200EI 205
>AI967395
Length = 205
Score = 71.6 bits (174), Expect = 6e-14
Identities = 32/66 (48%), Positives = 49/66 (73%)
Frame = +3
Query: 70 IEEAKLSVIVFSKNYAVSKWCLEELMKILEIKKMKGQIVVPVFYDVDPSDVRNQRGSYAE 129
I+E++L ++VFS++YAVS CL EL+ I+++ M Q+V PV+Y V+ ++R+QRG Y E
Sbjct: 3 IQESRLIIVVFSEHYAVSASCLNELVNIVDVMIMNNQLVWPVYYGVEAPEIRHQRGRYGE 182
Query: 130 AFAKHE 135
A AK E
Sbjct: 183 AMAKFE 200
>TC12592
Length = 534
Score = 53.9 bits (128), Expect = 1e-08
Identities = 24/61 (39%), Positives = 39/61 (63%), Gaps = 1/61 (1%)
Frame = +3
Query: 106 QIVVPVFYDVDPSDVRNQRGSYAEAFAKHENNF-EGKIKVQEWRNGLLEAANYAGWDCNV 164
Q+V+PVFY V+P+D+RN +G + EA A+ EN F + + EW+ L + + G C+V
Sbjct: 3 QLVLPVFYGVEPTDIRNLKGKFGEAMAEIENKFGKDSNTIHEWKQALRDISYLKGEPCHV 182
Query: 165 N 165
+
Sbjct: 183 S 185
>TC15876 weakly similar to UP|Q8S4X3 (Q8S4X3) TIR-similar-domain-containing
protein TSDC, partial (13%)
Length = 558
Score = 53.1 bits (126), Expect = 2e-08
Identities = 26/61 (42%), Positives = 39/61 (63%), Gaps = 1/61 (1%)
Frame = +1
Query: 97 ILEIKKMKGQIVVPVFYDVDPSDVRNQRGSYAEAFAKHENNF-EGKIKVQEWRNGLLEAA 155
ILE + Q+V P+FY V+PSDVR+QR SY +A K E + + KV++WR+ L +
Sbjct: 16 ILECXQKNDQLVWPIFYKVEPSDVRHQRNSYEKAMIKPETRYGKDSEKVKKWRSSLSQVC 195
Query: 156 N 156
+
Sbjct: 196 D 198
>TC20188 weakly similar to UP|Q8S4X3 (Q8S4X3) TIR-similar-domain-containing
protein TSDC, partial (11%)
Length = 494
Score = 50.4 bits (119), Expect = 2e-07
Identities = 25/55 (45%), Positives = 34/55 (61%), Gaps = 1/55 (1%)
Frame = +1
Query: 106 QIVVPVFYDVDPSDVRNQRGSYAEAFAKHENNF-EGKIKVQEWRNGLLEAANYAG 159
Q+V P+FY++ SDV NQ SY A HEN+F + KVQ+W++ L E A G
Sbjct: 4 QLVWPIFYEISXSDVSNQTKSYGHAMLAHENSFGKDSEKVQKWKSALSEMAFLQG 168
>AV408271
Length = 409
Score = 50.1 bits (118), Expect = 2e-07
Identities = 22/45 (48%), Positives = 31/45 (68%)
Frame = +1
Query: 18 EKHEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDNDLNSGDEI 62
EK++VF+SFRGED R+ F SHL A ++ I ++D+ L G EI
Sbjct: 274 EKYDVFVSFRGEDIRDGFLSHLADAFRQKKINAFMDDKLKRGQEI 408
>BP068235
Length = 511
Score = 44.7 bits (104), Expect(2) = 3e-07
Identities = 25/63 (39%), Positives = 35/63 (54%), Gaps = 2/63 (3%)
Frame = -1
Query: 91 LEELMKILEIKKMKGQIVVPVFYDVDPSDVRNQRGSYAEAFAKHENNFEG--KIKVQEWR 148
L L + LE KKMK Q++ P Y V+PSD+R + SY A A + G K+Q W+
Sbjct: 475 LMNLSRFLECKKMKNQLLWPCPYKVEPSDIRYRPSSYGTALA*TRTDAFGNDSEKIQTWK 296
Query: 149 NGL 151
+ L
Sbjct: 295 SAL 287
Score = 24.3 bits (51), Expect(2) = 3e-07
Identities = 10/17 (58%), Positives = 14/17 (81%)
Frame = -3
Query: 81 SKNYAVSKWCLEELMKI 97
S+NYA S CL+EL++I
Sbjct: 506 SENYAYSARCLDELVQI 456
>TC9932 weakly similar to UP|Q93YA7 (Q93YA7) Resistance gene-like, partial
(5%)
Length = 689
Score = 46.2 bits (108), Expect = 3e-06
Identities = 27/66 (40%), Positives = 42/66 (62%), Gaps = 1/66 (1%)
Frame = -2
Query: 6 SSSSSSFNNAPLEKHEVFISFRGEDTRNNFTSHLNGALKRLDIRTYIDND-LNSGDEIPT 64
S SSSS N+ + ++VFI+FRG+DTR++F SHL AL + ++D++ L G E+
Sbjct: 238 SMSSSSSNHQWI--YDVFINFRGKDTRHSFVSHLYTALSNAGVNVFLDDENLERGLELEP 65
Query: 65 TLVRAI 70
L + I
Sbjct: 64 ELQKLI 47
>BP070696
Length = 429
Score = 33.5 bits (75), Expect = 0.019
Identities = 17/55 (30%), Positives = 30/55 (53%), Gaps = 1/55 (1%)
Frame = -1
Query: 98 LEIKKMKGQIVVPVFYDVDPSDVRNQRGSYAEAFAKHE-NNFEGKIKVQEWRNGL 151
LE KK K I+ P+F +++ + Q SY + HE N + +V++W++ L
Sbjct: 429 LECKKKKNNIIRPIFQNMELLKIIYQINSYDDVMVAHERKNGKESDRVKKWKSAL 265
>BP083121
Length = 527
Score = 27.7 bits (60), Expect = 1.1
Identities = 12/24 (50%), Positives = 16/24 (66%)
Frame = -1
Query: 116 DPSDVRNQRGSYAEAFAKHENNFE 139
DP+DVR+Q+ SY +A K FE
Sbjct: 527 DPTDVRHQKNSYGQARKKVWKRFE 456
>TC17957 weakly similar to GB|AAM98310.1|22655436|AY142046
At2g19080/T20K24.9 {Arabidopsis thaliana;}, partial
(10%)
Length = 648
Score = 27.3 bits (59), Expect = 1.4
Identities = 14/32 (43%), Positives = 19/32 (58%)
Frame = -2
Query: 17 LEKHEVFISFRGEDTRNNFTSHLNGALKRLDI 48
L K E I RGE+ RN + HLN K+L++
Sbjct: 644 LGKLESSIKTRGENVRNLYHPHLN*CTKKLNL 549
>TC12438 similar to UP|ALG6_ARATH (Q9FF17) Probable dolichyl pyrophosphate
Man9GlcNAc2 alpha-1,3-glucosyltransferase
(Dolichyl-P-Glc:Man9GlcNAc2-PP-dolichyl
glucosyltransferase) , partial (23%)
Length = 491
Score = 26.9 bits (58), Expect = 1.8
Identities = 15/52 (28%), Positives = 25/52 (47%)
Frame = +3
Query: 96 KILEIKKMKGQIVVPVFYDVDPSDVRNQRGSYAEAFAKHENNFEGKIKVQEW 147
+++ + +G + P+F PS+VR RG AEA + VQ+W
Sbjct: 165 RVVRVAGPRGGVAAPLFRRRKPSEVRGLRG--AEALDGDNHQPSDSRMVQKW 314
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.132 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,722,418
Number of Sequences: 28460
Number of extensions: 30338
Number of successful extensions: 161
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 156
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 156
length of query: 166
length of database: 4,897,600
effective HSP length: 83
effective length of query: 83
effective length of database: 2,535,420
effective search space: 210439860
effective search space used: 210439860
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 52 (24.6 bits)
Medicago: description of AC134049.16


