

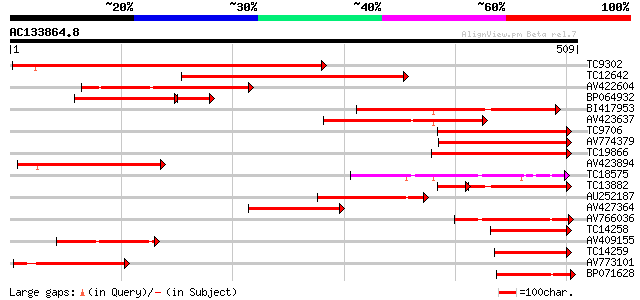
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133864.8 - phase: 0
(509 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC9302 similar to UP|BGLT_TRIRP (P26205) Cyanogenic beta-glucosi... 348 1e-96
TC12642 weakly similar to UP|Q7X9A9 (Q7X9A9) Beta-primeverosidas... 253 5e-68
AV422604 205 1e-53
BP064932 125 2e-40
BI417953 160 4e-40
AV423637 158 2e-39
TC9706 similar to UP|Q8W594 (Q8W594) Prunasin hydrolase isoform ... 139 1e-33
AV774379 136 7e-33
TC19866 similar to UP|Q8L7J2 (Q8L7J2) Beta-glucosidase, partial ... 136 9e-33
AV423894 132 1e-31
TC18575 similar to GB|BAB02020.1|9294063|AB020749 beta-glucosida... 121 3e-28
TC13882 weakly similar to UP|Q8W594 (Q8W594) Prunasin hydrolase ... 95 1e-25
AU252187 102 1e-22
AV427364 101 3e-22
AV766036 98 4e-21
TC14258 similar to UP|Q945I3 (Q945I3) Prunasin hydrolase isoform... 94 4e-20
AV409155 92 3e-19
TC14259 similar to UP|Q43073 (Q43073) Prunasin hydrolase precurs... 91 4e-19
AV773101 89 2e-18
BP071628 76 1e-14
>TC9302 similar to UP|BGLT_TRIRP (P26205) Cyanogenic beta-glucosidase
precursor (Linamarase) (Fragment) , partial (58%)
Length = 867
Score = 348 bits (893), Expect = 1e-96
Identities = 165/285 (57%), Positives = 209/285 (72%), Gaps = 3/285 (1%)
Frame = +2
Query: 3 AISHFLLYLFSLATLLAVV--TGTASQHVHPSHYAASFNRTLFPSDFLFGIGSSAYQIEG 60
A++ FL++L L LL + T +Q V P AS NR+ FP F+FG SSAYQ EG
Sbjct: 11 ALNTFLVFLLPLLILLVCTLPSFTFAQEVSPIVDVASLNRSSFPKGFIFGTASSAYQYEG 190
Query: 61 ASNIDGRGPSIWDTFTKQHPEKIGDHSSGNIGADFYHRYKSDIKIMKEIGLDSYRFSISW 120
A+N GR PSIWDT+ + E+I D S+G++ D YHRYK D+ IMK + +D+YRFSISW
Sbjct: 191 AANKYGRKPSIWDTYAHNYSERIVDRSNGDVAVDEYHRYKEDVGIMKSMNMDAYRFSISW 370
Query: 121 SRIFPSKKGKGAVNPMGVKFYNNVINEVLANGLIPFVTLFHWDLPQSLEDEYKGFLSPKI 180
SRI P K +G +N G+K+YNN+INE+LANGL P+VTLFHWD+PQ+LEDEY GFLSP +
Sbjct: 371 SRILPKGKLRGGINQEGIKYYNNLINELLANGLQPYVTLFHWDMPQALEDEYGGFLSPHV 550
Query: 181 VKDFEAYADFCFKTFGDRVKHWVTLNEPVSYTINGYHGGTSPPARCSKYVG-NCSTGDST 239
VKDF YA+ CFK FGDRVKHW+TLNEP YT NGY G P RCSK++ NC+ GDS
Sbjct: 551 VKDFRDYAELCFKEFGDRVKHWITLNEPWGYTSNGYALGEFAPGRCSKWLDPNCTGGDSG 730
Query: 240 TEPYIVAHHFILSHAAAAKLYKAKYQAHQKGKIGITLITHYYEPY 284
TEPY+V H+ +L+HA A +YK KYQA QKG IG+TL+TH++EP+
Sbjct: 731 TEPYLVTHYQLLAHAEAVHVYKKKYQASQKGIIGVTLVTHWFEPF 865
>TC12642 weakly similar to UP|Q7X9A9 (Q7X9A9) Beta-primeverosidase ,
partial (40%)
Length = 652
Score = 253 bits (646), Expect = 5e-68
Identities = 115/204 (56%), Positives = 151/204 (73%)
Frame = +3
Query: 155 PFVTLFHWDLPQSLEDEYKGFLSPKIVKDFEAYADFCFKTFGDRVKHWVTLNEPVSYTIN 214
PF+TLFH DLPQ+LEDEY GFL+PK +DF YA+ CF+ FGDRVKH +TLNEP +
Sbjct: 18 PFITLFHSDLPQALEDEYGGFLNPKXEQDFADYAEVCFREFGDRVKHXITLNEPALTSTQ 197
Query: 215 GYHGGTSPPARCSKYVGNCSTGDSTTEPYIVAHHFILSHAAAAKLYKAKYQAHQKGKIGI 274
GY G SPP RCSK+V NCS G+S TEPYIV H+ IL+HAAA K+Y+ K+Q QKG+IGI
Sbjct: 198 GYGNGGSPPLRCSKWVANCSAGNSATEPYIVTHNLILAHAAAVKVYREKFQISQKGQIGI 377
Query: 275 TLITHYYEPYSNSVADHKAASRALDFLFGWFAHPITYGHYPQSMISSLGNRLPKFTKEEY 334
TL + + P S S D +AASR L F++ WF P+ G YP M++ +G RLP+FT+ +
Sbjct: 378 TLNSAWVVPLSQSKDDIEAASRGLAFMYDWFMEPLNSGKYPAVMVNRVGKRLPEFTRSQS 557
Query: 335 KIIKGSYDFLGVNYYTTYYAQSIP 358
++KGS+DF+G+NYYT+ YA ++P
Sbjct: 558 LMVKGSFDFIGLNYYTSTYAANVP 629
>AV422604
Length = 460
Score = 205 bits (522), Expect = 1e-53
Identities = 90/155 (58%), Positives = 121/155 (78%)
Frame = +1
Query: 65 DGRGPSIWDTFTKQHPEKIGDHSSGNIGADFYHRYKSDIKIMKEIGLDSYRFSISWSRIF 124
DGRGPS+WDTF+ KI DHS+ ++ D YHRY+ DI++MK++G+D+YRFSISW+RIF
Sbjct: 1 DGRGPSVWDTFSHTFG-KITDHSNADVAVDQYHRYEEDIQLMKDLGMDAYRFSISWTRIF 177
Query: 125 PSKKGKGAVNPMGVKFYNNVINEVLANGLIPFVTLFHWDLPQSLEDEYKGFLSPKIVKDF 184
P+ G G +N GV YN +IN +LA G+ P+VT++HWDLPQ+LED+Y G+LSP+I+KDF
Sbjct: 178 PN--GSGEINQAGVDHYNKLINALLAKGVEPYVTMYHWDLPQALEDKYNGWLSPEIIKDF 351
Query: 185 EAYADFCFKTFGDRVKHWVTLNEPVSYTINGYHGG 219
YA+ CF+ FGDRVKHW+T NEP ++T GY G
Sbjct: 352 ANYAETCFQKFGDRVKHWITFNEPHTFTTQGYDVG 456
>BP064932
Length = 459
Score = 125 bits (313), Expect(2) = 2e-40
Identities = 56/94 (59%), Positives = 71/94 (74%)
Frame = +2
Query: 59 EGASNIDGRGPSIWDTFTKQHPEKIGDHSSGNIGADFYHRYKSDIKIMKEIGLDSYRFSI 118
EGA+N GRGPSIWDTFT ++PE I D S+G++ D YHRYK D++IMK + D+YRFSI
Sbjct: 11 EGAANEGGRGPSIWDTFTHKNPETILDRSNGDVAVDQYHRYKEDVQIMKYMNTDAYRFSI 190
Query: 119 SWSRIFPSKKGKGAVNPMGVKFYNNVINEVLANG 152
SWSRI P K +N G+K+YNN+INE+L NG
Sbjct: 191 SWSRILPKGKISAGINQEGIKYYNNLINELLDNG 292
Score = 57.8 bits (138), Expect(2) = 2e-40
Identities = 24/34 (70%), Positives = 28/34 (81%)
Frame = +1
Query: 151 NGLIPFVTLFHWDLPQSLEDEYKGFLSPKIVKDF 184
+GL PFVTLFHWDLPQ+L+DEY GF SP I+ F
Sbjct: 358 SGLQPFVTLFHWDLPQALQDEYSGFSSPHIINGF 459
>BI417953
Length = 564
Score = 160 bits (406), Expect = 4e-40
Identities = 76/189 (40%), Positives = 116/189 (61%), Gaps = 6/189 (3%)
Frame = +2
Query: 312 GHYPQSMISSLGNRLPKFTKEEYKIIKGSYDFLGVNYYTTYYAQSIPPTYINMTYFTDMQ 371
G YP M+S LG RLPKF++E+ ++K S+DF+G+NYY+T YA N TY TD
Sbjct: 8 GSYPVEMVSYLGERLPKFSEEQSAMLKNSFDFIGINYYSTAYAADAECPSQNRTYLTDYC 187
Query: 372 ANLIHSNE------EWSNYWLLVYPKGIHHLVTHIKDTYKNPPVYITENGIGQSRNDSIP 425
A L + + ++ W+ +YP+GI ++ + K+ + NP +YITENG + +
Sbjct: 188 AKLTYERDGVPIGPRAASVWIYLYPRGIEEVMLYFKNKFNNPVMYITENGYDEFNDGKGS 367
Query: 426 VNVARKDGIRIRYHDSHLKFLLQAIKDGANVKGYYAWSFSDSYEWDAGYTVRFGIIYVDF 485
++ D RI H HL ++ AI G +V+GY+ WS D++EW GYTVRFG++YV++
Sbjct: 368 LH----DQERIDCHIQHLSYVHSAILKGVDVRGYFVWSLLDNFEWSDGYTVRFGLVYVNY 535
Query: 486 VNNLKRYPK 494
+ L+RYPK
Sbjct: 536 TDGLQRYPK 562
>AV423637
Length = 469
Score = 158 bits (400), Expect = 2e-39
Identities = 73/154 (47%), Positives = 104/154 (67%), Gaps = 6/154 (3%)
Frame = +3
Query: 282 EPYSNSVADHKAASRALDFLFGWFAHPITYGHYPQSMISSLGNRLPKFTKEEYKIIKGSY 341
EP+S++ DH AA RA+DF+FGW+ P+T+G YP SMIS +GNRLPKFT + +++KGS+
Sbjct: 3 EPFSDNKYDHHAAGRAIDFMFGWYMDPLTFGKYPDSMISLVGNRLPKFTSRQARLVKGSF 182
Query: 342 DFLGVNYYTTYYAQSIPPTYINMTYFTDMQANLIHSNE------EWSNYWLLVYPKGIHH 395
DF+G+NYYTTYYA + PP I+ +FTD ANL ++ WL +YPKGI
Sbjct: 183 DFIGINYYTTYYAANAPPG-IHPYFFTDSLANLTGERNGNPIGPRAASTWLYIYPKGIQE 359
Query: 396 LVTHIKDTYKNPPVYITENGIGQSRNDSIPVNVA 429
L+ + K Y NP +YITENG+ + + ++ + A
Sbjct: 360 LLLYTKKKYNNPLIYITENGMSEFNDPTLSLEEA 461
>TC9706 similar to UP|Q8W594 (Q8W594) Prunasin hydrolase isoform PH C
precursor , partial (24%)
Length = 677
Score = 139 bits (350), Expect = 1e-33
Identities = 64/120 (53%), Positives = 83/120 (68%)
Frame = +3
Query: 385 WLLVYPKGIHHLVTHIKDTYKNPPVYITENGIGQSRNDSIPVNVARKDGIRIRYHDSHLK 444
WL VYPKGI L+ +IK Y NP +YITENG+ + ++ + A D RI Y+ HL
Sbjct: 48 WLYVYPKGIQELLLYIKKKYNNPLIYITENGMSDFNDPTLSLEEALLDTFRIDYYYRHLF 227
Query: 445 FLLQAIKDGANVKGYYAWSFSDSYEWDAGYTVRFGIIYVDFVNNLKRYPKYSAFWLQKFL 504
+L AI+DGANVKGY+AWS D++EW +GYT+RFGI +VD+ N LKRY K SA W + FL
Sbjct: 228 YLQSAIRDGANVKGYFAWSLLDNFEWASGYTLRFGINFVDYKNGLKRYQKLSAKWFKNFL 407
>AV774379
Length = 455
Score = 136 bits (343), Expect = 7e-33
Identities = 63/119 (52%), Positives = 83/119 (68%)
Frame = -1
Query: 386 LLVYPKGIHHLVTHIKDTYKNPPVYITENGIGQSRNDSIPVNVARKDGIRIRYHDSHLKF 445
L VYPKGI L+ ++K Y NP +YIT NGI + + ++ + A D RI Y+ HL +
Sbjct: 455 LYVYPKGIRELLLYVKKKYNNPQIYITVNGIDEFNDPTLSLEEALLDTFRIDYYYRHLFY 276
Query: 446 LLQAIKDGANVKGYYAWSFSDSYEWDAGYTVRFGIIYVDFVNNLKRYPKYSAFWLQKFL 504
L AI+DGANVKGY+AWS D++EW +GYTVRFGI +VD+ N +KRYPK SA W + FL
Sbjct: 275 LQSAIRDGANVKGYFAWSLLDNFEWFSGYTVRFGINFVDYKNGMKRYPKLSAKWFRYFL 99
>TC19866 similar to UP|Q8L7J2 (Q8L7J2) Beta-glucosidase, partial (25%)
Length = 562
Score = 136 bits (342), Expect = 9e-33
Identities = 63/126 (50%), Positives = 84/126 (66%)
Frame = +1
Query: 379 EEWSNYWLLVYPKGIHHLVTHIKDTYKNPPVYITENGIGQSRNDSIPVNVARKDGIRIRY 438
E ++ WL + P+G+ L+ +IK Y NP V ITENG+ N I + A KD RIRY
Sbjct: 25 ERANSIWLYLVPEGMRSLMNYIKQKYGNPLVVITENGMDDPNNPFISIKDALKDEKRIRY 204
Query: 439 HDSHLKFLLQAIKDGANVKGYYAWSFSDSYEWDAGYTVRFGIIYVDFVNNLKRYPKYSAF 498
++ +L LL +IKDG NVKGY+AWS D++EW AGYT RFG+ +VD+ +N KRYPK S
Sbjct: 205 YNGYLSSLLASIKDGCNVKGYFAWSLLDNWEWSAGYTCRFGLYFVDYKDNQKRYPKQSVE 384
Query: 499 WLQKFL 504
W + FL
Sbjct: 385 WFKNFL 402
>AV423894
Length = 408
Score = 132 bits (333), Expect = 1e-31
Identities = 65/135 (48%), Positives = 85/135 (62%), Gaps = 2/135 (1%)
Frame = +2
Query: 8 LLYLFSLATLLAVVTG--TASQHVHPSHYAASFNRTLFPSDFLFGIGSSAYQIEGASNID 65
L++L L +LL T ++ V P S NR+ FP F+FG SSAYQ EGA+N
Sbjct: 2 LVFLLPLLSLLVCTLPSITFAEDVSPIADVVSLNRSSFPKGFIFGTASSAYQYEGAANKG 181
Query: 66 GRGPSIWDTFTKQHPEKIGDHSSGNIGADFYHRYKSDIKIMKEIGLDSYRFSISWSRIFP 125
GR PSIWDT+ H ++I D S+G++ D YHRYK D+ IMK + LD+YRFSISW RI P
Sbjct: 182 GRKPSIWDTYAHNHSDRIADGSNGDVAIDEYHRYKEDVGIMKSMNLDAYRFSISWPRILP 361
Query: 126 SKKGKGAVNPMGVKF 140
K G +N G+K+
Sbjct: 362 KGKLSGGINQEGIKY 406
>TC18575 similar to GB|BAB02020.1|9294063|AB020749 beta-glucosidase
{Arabidopsis thaliana;} , partial (35%)
Length = 797
Score = 121 bits (303), Expect = 3e-28
Identities = 80/210 (38%), Positives = 113/210 (53%), Gaps = 14/210 (6%)
Frame = +1
Query: 307 HPITYGHYPQSMISSLGNRLPKFTKEEYKIIKGSYDFLGVNYYTTYYA----QSIPPTYI 362
HP+ G YP+++ + +GNRLPKFTKEE KI+KGS+DF+G+N YTTYY QS P
Sbjct: 16 HPLVCGEYPKTIQNIVGNRLPKFTKEEVKIVKGSFDFIGINQYTTYYMYDPHQSKPKV-- 189
Query: 363 NMTYFTDMQANLIHSNE------EWSNYWLLVYPKGIHHLVTHIKDTYKNPPVYITENGI 416
Y + A ++ + +YWL P G++ + +IK+ Y NP V ++ENG+
Sbjct: 190 -PGYQQEWNAGFAYAKNGVPIGPQAYSYWLYNVPWGMYKALMYIKERYGNPTVILSENGM 366
Query: 417 GQSRNDSIPVNVARKDGIRIRYHDSHLKFLLQAIKDGANVKG----YYAWSFSDSYEWDA 472
N I A D RI Y+ ++L L +A DGANV G + W + S +W
Sbjct: 367 DDPGN--ITFT*ALHDTTRIYYY*TYLTQLKKARDDGANVVG*CSHGHCW-ITSSGDW-- 531
Query: 473 GYTVRFGIIYVDFVNNLKRYPKYSAFWLQK 502
G + DF L+R PK SAFW +K
Sbjct: 532 GTHQGLALCMFDF-KTLQRTPKMSAFWFKK 618
>TC13882 weakly similar to UP|Q8W594 (Q8W594) Prunasin hydrolase isoform PH
C precursor , partial (23%)
Length = 631
Score = 95.1 bits (235), Expect(2) = 1e-25
Identities = 44/92 (47%), Positives = 63/92 (67%)
Frame = +1
Query: 413 ENGIGQSRNDSIPVNVARKDGIRIRYHDSHLKFLLQAIKDGANVKGYYAWSFSDSYEWDA 472
+NGI + + + ++ D +RI Y HL +L +AI++G VKGY+AWS D++EW A
Sbjct: 202 KNGIDEVNDGKMSLD----DKVRIDYILRHLLYLQRAIRNGVRVKGYFAWSLLDNFEWTA 369
Query: 473 GYTVRFGIIYVDFVNNLKRYPKYSAFWLQKFL 504
GYT+RFG++YVDF N L+RY K SA W + FL
Sbjct: 370 GYTLRFGLVYVDFKNGLRRYHKRSALWFKLFL 465
Score = 38.1 bits (87), Expect(2) = 1e-25
Identities = 15/29 (51%), Positives = 21/29 (71%)
Frame = +3
Query: 385 WLLVYPKGIHHLVTHIKDTYKNPPVYITE 413
WL VYP+GI L+ + K+ + NP V+ITE
Sbjct: 117 WLYVYPRGIQGLLEYTKEKFNNPIVFITE 203
>AU252187
Length = 350
Score = 102 bits (255), Expect = 1e-22
Identities = 47/100 (47%), Positives = 69/100 (69%)
Frame = +1
Query: 277 ITHYYEPYSNSVADHKAASRALDFLFGWFAHPITYGHYPQSMISSLGNRLPKFTKEEYKI 336
+T+++EP ++ DH AA RA+DF+ GW +P+T G YPQSM S +GNRLP+F+ ++ ++
Sbjct: 1 VTYWFEPLLDNKYDHDAAGRAIDFMLGWHLNPLTTGKYPQSMRSLVGNRLPEFSLKQARL 180
Query: 337 IKGSYDFLGVNYYTTYYAQSIPPTYINMTYFTDMQANLIH 376
I GS+DF+G+NYYTTYYA + + TD A L H
Sbjct: 181 INGSFDFIGLNYYTTYYATN-ASSVSQPNSITDSLAYLTH 297
>AV427364
Length = 268
Score = 101 bits (252), Expect = 3e-22
Identities = 49/86 (56%), Positives = 59/86 (67%)
Frame = +1
Query: 215 GYHGGTSPPARCSKYVGNCSTGDSTTEPYIVAHHFILSHAAAAKLYKAKYQAHQKGKIGI 274
GY G P RCSK GNC+ G+S TEPYIV H+ +LSHAAA + Y+ KYQ QKG+IGI
Sbjct: 4 GYDNGFFAPGRCSKEYGNCTAGNSGTEPYIVTHNLLLSHAAAVQRYREKYQEKQKGRIGI 183
Query: 275 TLITHYYEPYSNSVADHKAASRALDF 300
L +YEP + S AD+ AA RA DF
Sbjct: 184 LLDFVWYEPLTRSKADNNAAQRARDF 261
>AV766036
Length = 518
Score = 97.8 bits (242), Expect = 4e-21
Identities = 52/107 (48%), Positives = 66/107 (61%)
Frame = -1
Query: 400 IKDTYKNPPVYITENGIGQSRNDSIPVNVARKDGIRIRYHDSHLKFLLQAIKDGANVKGY 459
IK+ Y +P V+ +ENG+ N I A RI Y+ ++L L +A DGANV GY
Sbjct: 509 IKERYGDPTVFFSENGMDGPGN--ITFTKALHGATRINYYKTYLPPLKKARDDGANVVGY 336
Query: 460 YAWSFSDSYEWDAGYTVRFGIIYVDFVNNLKRYPKYSAFWLQKFLLK 506
+AWS D++EW GY RFGI+YVDF LKR PK SAFW +K L K
Sbjct: 335 FAWSLLDNFEWRLGYPSRFGIVYVDF-KTLKRTPKMSAFWFKKLLSK 198
>TC14258 similar to UP|Q945I3 (Q945I3) Prunasin hydrolase isoform PH A
(Fragment) , partial (15%)
Length = 602
Score = 94.4 bits (233), Expect = 4e-20
Identities = 42/73 (57%), Positives = 53/73 (72%)
Frame = +2
Query: 432 DGIRIRYHDSHLKFLLQAIKDGANVKGYYAWSFSDSYEWDAGYTVRFGIIYVDFVNNLKR 491
D RI Y+ HL +L AI++G+NVKGY+AWS D+YEW +GYTVRFG+ +VD+ N LKR
Sbjct: 2 DTFRIDYYFRHLFYLQSAIRNGSNVKGYFAWSLLDNYEWSSGYTVRFGMNFVDYENGLKR 181
Query: 492 YPKYSAFWLQKFL 504
Y K SA W FL
Sbjct: 182 YKKLSAKWFTNFL 220
>AV409155
Length = 426
Score = 91.7 bits (226), Expect = 3e-19
Identities = 47/93 (50%), Positives = 64/93 (68%), Gaps = 1/93 (1%)
Frame = +3
Query: 43 FPSDFLFGIGSSAYQIEGASNIDGRGPSIWDTFTKQHPEKIGDHSSGNIGADFYHRYKSD 102
F +FLFG SS+YQ EGA DG+G + WD FT + P D ++G+I D YHRY+ D
Sbjct: 153 FSDNFLFGTASSSYQFEGAYLTDGKGLNNWDVFTHK-PGTTMDGTNGDIAVDHYHRYEED 329
Query: 103 IKIMKEIGLDSYRFSISWSRIFPSKKGK-GAVN 134
+ +M+ IG++SYRFS+SW+RI P KG+ G VN
Sbjct: 330 VDLMEFIGVNSYRFSLSWARILP--KGRYGKVN 422
>TC14259 similar to UP|Q43073 (Q43073) Prunasin hydrolase precursor ,
partial (13%)
Length = 568
Score = 90.9 bits (224), Expect = 4e-19
Identities = 40/69 (57%), Positives = 51/69 (72%)
Frame = +3
Query: 436 IRYHDSHLKFLLQAIKDGANVKGYYAWSFSDSYEWDAGYTVRFGIIYVDFVNNLKRYPKY 495
I Y+ HL +L AI++G+NVKGY+AWS D+YEW +GYTVRFG+ +VD+ N LKRY K
Sbjct: 3 IDYYFRHLFYLRSAIRNGSNVKGYFAWSLLDNYEWSSGYTVRFGMNFVDYKNGLKRYKKL 182
Query: 496 SAFWLQKFL 504
SA W FL
Sbjct: 183 SAKWFTNFL 209
>AV773101
Length = 526
Score = 88.6 bits (218), Expect = 2e-18
Identities = 46/105 (43%), Positives = 64/105 (60%), Gaps = 1/105 (0%)
Frame = +1
Query: 4 ISHFLLYLFSLATLLAVVTGTASQHV-HPSHYAASFNRTLFPSDFLFGIGSSAYQIEGAS 62
+ + L+ LFSL T+S HV P + NR+ FP F++G S+AYQ EG +
Sbjct: 229 VMNVLVCLFSLLA-------TSSAHVVAPILDDSDLNRSSFPPGFVYGTASAAYQHEGGA 387
Query: 63 NIDGRGPSIWDTFTKQHPEKIGDHSSGNIGADFYHRYKSDIKIMK 107
G+GPS+WDTFT +HPEKI ++ ++ D YHRYK D+ IMK
Sbjct: 388 REGGKGPSVWDTFTHKHPEKIAGGANADVTCDQYHRYKEDVGIMK 522
>BP071628
Length = 402
Score = 76.3 bits (186), Expect = 1e-14
Identities = 32/71 (45%), Positives = 50/71 (70%)
Frame = -1
Query: 438 YHDSHLKFLLQAIKDGANVKGYYAWSFSDSYEWDAGYTVRFGIIYVDFVNNLKRYPKYSA 497
Y HL L++AI+ GA+V+GY+ WS D++EW GYT+RFG+ +VD+ +KR P+ SA
Sbjct: 333 YMAGHLDALMEAIRKGADVRGYFGWSLLDNFEWLRGYTIRFGLHHVDYA-TMKRTPRLSA 157
Query: 498 FWLQKFLLKGK 508
W ++F+ + K
Sbjct: 156 IWYKEFIARHK 124
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.321 0.138 0.434
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,310,994
Number of Sequences: 28460
Number of extensions: 155590
Number of successful extensions: 737
Number of sequences better than 10.0: 55
Number of HSP's better than 10.0 without gapping: 715
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 723
length of query: 509
length of database: 4,897,600
effective HSP length: 94
effective length of query: 415
effective length of database: 2,222,360
effective search space: 922279400
effective search space used: 922279400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Medicago: description of AC133864.8


