

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133863.3 + phase: 0
(1104 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
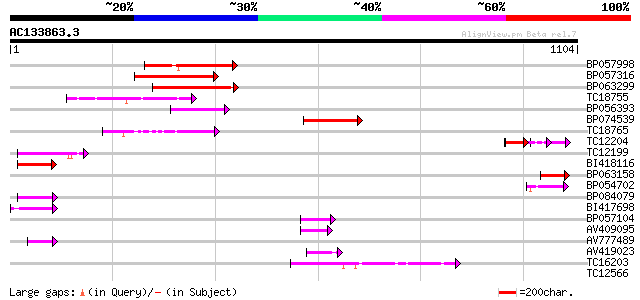
Score E
Sequences producing significant alignments: (bits) Value
BP057998 166 2e-41
BP057316 136 2e-32
BP063299 116 2e-26
TC18755 similar to UP|AAN85378 (AAN85378) Resistance protein (Fr... 115 3e-26
BP056393 101 7e-22
BP074539 94 1e-19
TC18765 weakly similar to UP|Q9LVT1 (Q9LVT1) Disease resistance ... 56 3e-08
TC12204 weakly similar to UP|Q9ZSN3 (Q9ZSN3) NBS-LRR-like protei... 55 6e-08
TC12199 53 2e-07
BI418116 53 2e-07
BP063158 53 3e-07
BP054702 49 4e-06
BP084079 48 1e-05
BI417698 45 5e-05
BP057104 44 1e-04
AV409095 44 1e-04
AV777489 44 1e-04
AV419023 44 1e-04
TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodula... 42 4e-04
TC12566 similar to UP|Q84XI1 (Q84XI1) RCa1 (Fragment), partial (... 41 0.001
>BP057998
Length = 566
Score = 166 bits (419), Expect = 2e-41
Identities = 91/187 (48%), Positives = 117/187 (61%), Gaps = 7/187 (3%)
Frame = +2
Query: 263 WEQLLLPFNHGSFGSKIIVTTREKEVAYHVVKSTMLCDLRQLVKSDCWRLFVTHAFQGKS 322
W QL PF HG GSKI+VTTR EVA VV++ +L QL DCW +F HA
Sbjct: 26 WNQLKKPFQHGMRGSKILVTTRSDEVA-SVVQTGQTFNLSQLSNEDCWSVFANHA----- 187
Query: 323 VCDYP-------KLESIGRKIMDKCEGLPLAIISLGQLLRKKFSQDEWMKILETDMWRLS 375
C +P LE IG +I+ KC+GLPLA SLG LLR+K + +W +L D+W LS
Sbjct: 188 -CLFPGLGENTIALEKIGLEIVKKCKGLPLAAQSLGGLLRRKHNIKDWNNVLNCDIWELS 364
Query: 376 DVDNKINPVLRLSYHNLPSDQKRCFAFCSIFPKGYTFEKDELIKLWMAEGLLKCCGSYKS 435
+ ++KI P LR+SYH LPS KRCFA+ S++PK Y FEK+++I LWMAE LL K+
Sbjct: 365 ESESKIIPSLRISYHYLPSYLKRCFAYFSLYPKDYEFEKNDVILLWMAEDLLPPPKQGKT 544
Query: 436 EEEFGNE 442
E G+E
Sbjct: 545 LNEVGDE 565
>BP057316
Length = 494
Score = 136 bits (343), Expect = 2e-32
Identities = 71/164 (43%), Positives = 100/164 (60%)
Frame = +2
Query: 243 LMGKKYLLVLDDIWNGNVEYWEQLLLPFNHGSFGSKIIVTTREKEVAYHVVKSTMLCDLR 302
L GKKYLLVLDD WN + + W QL G+ GSK++VTTR + VA + ++ L
Sbjct: 5 LSGKKYLLVLDDTWNESYQKWTQLRKYLMCGAQGSKVLVTTRSQIVA-QTMGVQVIHVLN 181
Query: 303 QLVKSDCWRLFVTHAFQGKSVCDYPKLESIGRKIMDKCEGLPLAIISLGQLLRKKFSQDE 362
L + W L F+ + LESIG KI +KC G+PLAI +LG LL+ K + E
Sbjct: 182 SLTLEESWGLLKKITFEDDATQVSQSLESIGEKIAEKCRGVPLAIRTLGGLLQSKSEESE 361
Query: 363 WMKILETDMWRLSDVDNKINPVLRLSYHNLPSDQKRCFAFCSIF 406
WM +L+ D W+L + ++ I PVL+LSY NL ++CFA+CS++
Sbjct: 362 WMNVLQGDFWKLCEDEDSIIPVLKLSYQNLSPQIRQCFAYCSLY 493
>BP063299
Length = 580
Score = 116 bits (290), Expect = 2e-26
Identities = 67/166 (40%), Positives = 102/166 (61%)
Frame = +3
Query: 279 IIVTTREKEVAYHVVKSTMLCDLRQLVKSDCWRLFVTHAFQGKSVCDYPKLESIGRKIMD 338
++V+TR+ +VA ++ + L+ L ++DCW LF +AF G + + +L +IG++I+
Sbjct: 54 VLVSTRDMDVA-EIMGTCQAQRLQGLSENDCWLLFKQYAF-GANNEERAELVAIGKEIVK 227
Query: 339 KCEGLPLAIISLGQLLRKKFSQDEWMKILETDMWRLSDVDNKINPVLRLSYHNLPSDQKR 398
KC GLPLA +LG LLR K EW ++ E+ +W L D + I P L LSY +L ++
Sbjct: 228 KCGGLPLAARALGGLLRLKNEVKEWHEVKESTLWNLPD-ERCILPALMLSYFHLTPTLRQ 404
Query: 399 CFAFCSIFPKGYTFEKDELIKLWMAEGLLKCCGSYKSEEEFGNEIF 444
CFAFC+IFPK K++LI LWMA G + + K E+ GN I+
Sbjct: 405 CFAFCAIFPKDAKIIKEDLIYLWMANGFISSTKN-KEVEDVGNSIW 539
>TC18755 similar to UP|AAN85378 (AAN85378) Resistance protein (Fragment),
complete
Length = 880
Score = 115 bits (289), Expect = 3e-26
Identities = 79/265 (29%), Positives = 134/265 (49%), Gaps = 12/265 (4%)
Frame = +3
Query: 111 LGLAEGPSASNEGLVSWKPSKRLSSTALVDESSIYGRDVDKEELIKFLLAGNDSGTQVPI 170
L A S+SN +W + L+D + + G D K++L+ L+ +
Sbjct: 102 LDTASEASSSNSSGNTWNDQR--GDALLLDNTDLVGIDRRKKKLMGCLIKPCPVRK---V 266
Query: 171 ISIVGLGGMGKTTLAKLVYNNNKIEEHFELKAWVYVSESYDVVGLTKAILKSFNP----- 225
IS+ G+GGMGKTTL K VY++ + +HF AW+ VS+S ++ L + + +
Sbjct: 267 ISVTGMGGMGKTTLVKQVYDDPVVIKHFRACAWITVSQSCEIGELLRDLARQLFSEIRRP 446
Query: 226 ---SADGEYLDQLQHQLQHMLMGKKYLLVLDDIWNGNVEYWEQLLLPFNHGSFGSKIIVT 282
+ D+L+ ++ +L ++YL+V DD+W +V WE + + GS+I++T
Sbjct: 447 VPLGLENMRCDRLKMIIKDLLQRRRYLVVFDDVW--HVREWEAVKYALPDNNCGSRIMIT 620
Query: 283 TREKEVAY--HVVKSTMLCDLRQLVKSDCWRLFVTHAFQGKSVCDYPKLESIGRKIMDKC 340
TR ++A+ + +L+ L + + W LF F G S + L I I+ KC
Sbjct: 621 TRRSDLAFTSSTESKGKVYNLQPLKEDEAWELFCRKTFHGDSCPSH--LIGICTYILRKC 794
Query: 341 EGLPLAIISLGQLLRKKFSQ--DEW 363
EGLPLAI+++ +L K + DEW
Sbjct: 795 EGLPLAIVAISGVLATKDKRRIDEW 869
>BP056393
Length = 421
Score = 101 bits (251), Expect = 7e-22
Identities = 52/116 (44%), Positives = 70/116 (59%)
Frame = +1
Query: 313 FVTHAFQGKSVCDYPKLESIGRKIMDKCEGLPLAIISLGQLLRKKFSQDEWMKILETDMW 372
F +HA E+IGR+I+ +C+G PL SLG LLR K + IL +++W
Sbjct: 64 FASHAGISLESTGNMTFETIGRQIVKRCKGSPLXAQSLGGLLRGKHDIRDXNNILNSNIW 243
Query: 373 RLSDVDNKINPVLRLSYHNLPSDQKRCFAFCSIFPKGYTFEKDELIKLWMAEGLLK 428
L + ++ I LR+SYH LP KRC +CS+FPK Y F+ DELI LWMAE LL+
Sbjct: 244 ELPENESNIILALRISYHYLPPHLKRCXVYCSLFPKDYKFDXDELILLWMAEDLLE 411
>BP074539
Length = 357
Score = 94.0 bits (232), Expect = 1e-19
Identities = 54/119 (45%), Positives = 76/119 (63%), Gaps = 3/119 (2%)
Frame = +1
Query: 572 NIKLLRYLDLSFTEITSLPDSICMLYNLQTILLQGCE-LTELPSNFSKLINLRHLEL--P 628
++K L YLDLS T I LPDS C+LYNLQ + L+ C L ELP N KL NLR+L+
Sbjct: 1 SLKHLCYLDLSHTNIEKLPDSTCLLYNLQILKLKNCRYLKELPLNLHKLTNLRYLDFSGT 180
Query: 629 YLKKMPKHIGKLNSLQTLPYFVVEEKNGSDLKELEKLNHLHGKICIDGLGYVFDPEDAV 687
++KMP H GKL +LQ L F V + + ++++L++LN L G + I L + +P DA+
Sbjct: 181 QVRKMPMHFGKLKNLQVLNSFFVGQGSEYNIQQLDELN-LQGTLSISELQNIINPLDAL 354
Score = 29.3 bits (64), Expect = 3.6
Identities = 19/57 (33%), Positives = 28/57 (48%)
Frame = +1
Query: 546 LFSRLNFLRTLSFRWCGLSELVDEISNIKLLRYLDLSFTEITSLPDSICMLYNLQTI 602
L L L+ + R+ L EL + + LRYLD S T++ +P L NLQ +
Sbjct: 70 LLYNLQILKLKNCRY--LKELPLNLHKLTNLRYLDFSGTQVRKMPMHFGKLKNLQVL 234
>TC18765 weakly similar to UP|Q9LVT1 (Q9LVT1) Disease resistance
protein-like, partial (18%)
Length = 688
Score = 56.2 bits (134), Expect = 3e-08
Identities = 66/239 (27%), Positives = 118/239 (48%), Gaps = 11/239 (4%)
Frame = +2
Query: 181 KTTLAKLVYNNNKIEEHF-ELKAWVYVSESYDVVGLTKAIL------KSFNPS-ADGEYL 232
KTTLA+ V ++++ HF E ++ VS+S +V L I + N + A +++
Sbjct: 2 KTTLAREVCRDDQVRCHFKERILFLTVSQSPNVEELRAKIFGHIMGNRGLNANYAVPQWM 181
Query: 233 DQLQHQLQHMLMGKKYLLVLDDIWNGNVEYWEQLLLPFNHGSFGSKIIVTTREKEVAYHV 292
Q + Q Q + L+VLDD+W ++ EQL+L G K +V +R K +
Sbjct: 182 PQFECQSQSQI-----LVVLDDVW--SLPVLEQLVLRVP----GCKYLVVSRFK--FQRI 322
Query: 293 VKSTMLCDLRQLVKSDCWRLFVTHAFQGKSVCDYPKLESIGRKIMDKCEGLPLAIISLGQ 352
T D+ L + D LF HAF KS+ + +++ ++++ +C LPLA+ +G
Sbjct: 323 FNDTY--DVELLSEGDALSLFCHHAFGHKSI-PFGANQNLIKQVVAECGRLPLALKVIGA 493
Query: 353 LLRKKFSQDEWMKILETDMWRLSDVDN-KINPV--LRLSYHNLPSDQKRCFAFCSIFPK 408
LR + ++ W+ + LS ++ ++N + + +S + LP K CF FP+
Sbjct: 494 SLRDQ-NEMFWLSVKTRLSQGLSIGESYEVNLIDRMAISTNYLPEKVKECFLDLCAFPE 667
>TC12204 weakly similar to UP|Q9ZSN3 (Q9ZSN3) NBS-LRR-like protein cD7
(Fragment), partial (3%)
Length = 328
Score = 55.1 bits (131), Expect = 6e-08
Identities = 34/78 (43%), Positives = 40/78 (50%)
Frame = +1
Query: 1015 LNSLKEFRVSDEFENVESFPEENLLPPNLRILLLYKCSKLRIMNYKGFLHLLSLSHLKIY 1074
L SL R+ + N ESFPE L PN+ L L KC KL+ + LLSL L I
Sbjct: 46 LQSLNSLRICN-CPNFESFPEGGLRAPNMTNLHLEKCKKLKSFPQQMNKMLLSLMTLNIK 222
Query: 1075 NCPSLERLPEKGLPKRRN 1092
CP LE +PE G P N
Sbjct: 223 ECPELESIPEGGFPDSLN 276
Score = 48.5 bits (114), Expect = 6e-06
Identities = 20/46 (43%), Positives = 28/46 (60%)
Frame = +1
Query: 965 LFTNLHSLYLYNCPELVSFPEGGLPSNLSCFSIFDCPKLIASREEW 1010
+ +L +L + CPEL S PEGG P +L+ IF C KL +R+ W
Sbjct: 190 MLLSLMTLNIKECPELESIPEGGFPDSLNLLEIFHCAKLFTNRKNW 327
Score = 45.8 bits (107), Expect = 4e-05
Identities = 32/93 (34%), Positives = 49/93 (52%), Gaps = 1/93 (1%)
Frame = +1
Query: 963 LYLFTNLHSLYLYNCPELVSFPEGGLPS-NLSCFSIFDCPKLIASREEWGLFQLNSLKEF 1021
L +L+SL + NCP SFPEGGL + N++ + C KL + ++ L SL
Sbjct: 37 LQYLQSLNSLRICNCPNFESFPEGGLRAPNMTNLHLEKCKKLKSFPQQMNKMLL-SLMTL 213
Query: 1022 RVSDEFENVESFPEENLLPPNLRILLLYKCSKL 1054
+ E +ES PE P +L +L ++ C+KL
Sbjct: 214 NIK-ECPELESIPEGG-FPDSLNLLEIFHCAKL 306
Score = 33.1 bits (74), Expect = 0.25
Identities = 27/95 (28%), Positives = 45/95 (46%), Gaps = 1/95 (1%)
Frame = +1
Query: 774 LGQLPSLRELSISNCKRIKIIGEELYGNNSKIDAFRSLEVLEFQRMENLEEWLCHEGFLS 833
L L SL L I NC + E + +L + + +++++ + + ++ LS
Sbjct: 37 LQYLQSLNSLRICNCPNFESFPE----GGLRAPNMTNLHLEKCKKLKSFPQQM-NKMLLS 201
Query: 834 LKELTIKDCPKLKRALPQHLP-SLQKLSIINCNKL 867
L L IK+CP+L+ P SL L I +C KL
Sbjct: 202 LMTLNIKECPELESIPEGGFPDSLNLLEIFHCAKL 306
>TC12199
Length = 551
Score = 53.1 bits (126), Expect = 2e-07
Identities = 45/156 (28%), Positives = 72/156 (45%), Gaps = 18/156 (11%)
Frame = +2
Query: 16 VIFEKLASVDIRDYFSSKNVDDLVKELNIALNSINHVLEEAEIKQYQIIYVKKWLDKLKH 75
V+FE LAS+ ++ + + + ++L+ L I V+E+AE KQ +K WL +LK
Sbjct: 86 VVFENLASLVQNEFATISGIKEKAEKLSQKLQVIKAVVEDAEKKQITNQSIKVWLQQLKD 265
Query: 76 VVYEADQLLDEISTDA-MLNKLKAESEPLTTNLLGVLG------------LAEGPS---- 118
Y D +LDE ST++ L L +P +G +AEG +
Sbjct: 266 AAYVLDDILDECSTESHRLAGLSTLIKPKNIMFRRKIGKKLKEITRRFDEIAEGKNKFIL 445
Query: 119 -ASNEGLVSWKPSKRLSSTALVDESSIYGRDVDKEE 153
S+ G P R + A + E +YGR +KE+
Sbjct: 446 QESDGGRRDEVPEGR-ETWATIHEPELYGRQEEKEK 550
>BI418116
Length = 584
Score = 53.1 bits (126), Expect = 2e-07
Identities = 27/75 (36%), Positives = 47/75 (62%)
Frame = +2
Query: 16 VIFEKLASVDIRDYFSSKNVDDLVKELNIALNSINHVLEEAEIKQYQIIYVKKWLDKLKH 75
+I EKL+S I+++ N+ D ++ + +++I VL +AE K WL++LK
Sbjct: 53 LILEKLSSSAIKEFGIIWNLKDDLQRMKNTVSAIKAVLRDAEAKATTNHQTSNWLEELKD 232
Query: 76 VVYEADQLLDEISTD 90
V+Y+AD LLD++ST+
Sbjct: 233 VLYDADDLLDDLSTE 277
Score = 28.9 bits (63), Expect = 4.6
Identities = 21/82 (25%), Positives = 39/82 (46%)
Frame = +2
Query: 682 DPEDAVTANLKDKKYLEELYMIFYDRKKEVDDSIVESNVSVLEALQPNRSLKRLSISQYR 741
D E T N + +LEEL + YD +DD E + + N+ +++L I +
Sbjct: 170 DAEAKATTNHQTSNWLEELKDVLYDADDLLDDLSTED--LKRQVMIGNKFVRKLQIFFSK 343
Query: 742 GNRFPNWIRGCHLPNLVSLQMR 763
N+ + ++ H N+ ++Q R
Sbjct: 344 SNQIAHGLKLGH--NMKAIQKR 403
>BP063158
Length = 431
Score = 52.8 bits (125), Expect = 3e-07
Identities = 25/57 (43%), Positives = 37/57 (64%)
Frame = -1
Query: 1033 FPEENLLPPNLRILLLYKCSKLRIMNYKGFLHLLSLSHLKIYNCPSLERLPEKGLPK 1089
FP+E LLP +L L + L+ ++Y+G HL SL+ L++ +C L LPE+GLPK
Sbjct: 431 FPDEGLLPHSLHYLDISHFPNLKKLDYRGLCHLPSLTELRLSDCSGLPCLPEEGLPK 261
Score = 31.2 bits (69), Expect = 0.94
Identities = 19/45 (42%), Positives = 23/45 (50%)
Frame = -1
Query: 753 HLPNLVSLQMRHCGLCSHLPPLGQLPSLRELSISNCKRIKIIGEE 797
H PNL L R GLC LPSL EL +S+C + + EE
Sbjct: 380 HFPNLKKLDYR--GLC-------HLPSLTELRLSDCSGLPCLPEE 273
Score = 31.2 bits (69), Expect = 0.94
Identities = 22/62 (35%), Positives = 31/62 (49%)
Frame = -1
Query: 724 EALQPNRSLKRLSISQYRGNRFPNWIRGCHLPNLVSLQMRHCGLCSHLPPLGQLPSLREL 783
E L P+ SL L IS + + ++ CHLP+L L++ C LP G S+ L
Sbjct: 422 EGLLPH-SLHYLDISHFPNLKKLDYRGLCHLPSLTELRLSDCSGLPCLPEEGLPKSISTL 246
Query: 784 SI 785
SI
Sbjct: 245 SI 240
>BP054702
Length = 528
Score = 48.9 bits (115), Expect = 4e-06
Identities = 34/88 (38%), Positives = 45/88 (50%), Gaps = 5/88 (5%)
Frame = -3
Query: 1006 SREEWG-----LFQLNSLKEFRVSDEFENVESFPEENLLPPNLRILLLYKCSKLRIMNYK 1060
S+ E+G L L+ L F D+ + + + +E LLP +L L LY S L+ +
Sbjct: 511 SKHEFGSQFPHLTSLSHLYVFGFGDQ-DLMNTLLKERLLPTSLVFLGLYNFSSLKFLEGN 335
Query: 1061 GFLHLLSLSHLKIYNCPSLERLPEKGLP 1088
G HL SL L I CPSL LPE LP
Sbjct: 334 GLQHLSSLHQLHIDKCPSLVSLPEDQLP 251
Score = 40.0 bits (92), Expect = 0.002
Identities = 22/53 (41%), Positives = 30/53 (56%), Gaps = 4/53 (7%)
Frame = -3
Query: 957 SSLSF----SLYLFTNLHSLYLYNCPELVSFPEGGLPSNLSCFSIFDCPKLIA 1005
SSL F L ++LH L++ CP LVS PE LP +L+ ++ CP L A
Sbjct: 361 SSLKFLEGNGLQHLSSLHQLHIDKCPSLVSLPEDQLPHSLAVLNLEKCPLLEA 203
>BP084079
Length = 508
Score = 47.8 bits (112), Expect = 1e-05
Identities = 27/77 (35%), Positives = 43/77 (55%)
Frame = +3
Query: 16 VIFEKLASVDIRDYFSSKNVDDLVKELNIALNSINHVLEEAEIKQYQIIYVKKWLDKLKH 75
V+FE L S+ ++ + + ++L+ L+ I VLE+AE KQ V WL +LK
Sbjct: 48 VVFENLLSLVQNEFATISGIKGKAEKLSHDLDLIKGVLEDAEKKQLTDRAVMVWLQQLKD 227
Query: 76 VVYEADQLLDEISTDAM 92
VY D +LDE S +++
Sbjct: 228 AVYVLDDILDECSIESL 278
>BI417698
Length = 519
Score = 45.4 bits (106), Expect = 5e-05
Identities = 29/92 (31%), Positives = 47/92 (50%)
Frame = +2
Query: 1 MAELVAGAFLQSSFQVIFEKLASVDIRDYFSSKNVDDLVKELNIALNSINHVLEEAEIKQ 60
MA+ + GA +F+KL S+ + + + ++L+ L I V+E+AE KQ
Sbjct: 89 MADALLGA--------VFDKLLSLAQNELATISGIKGKAQKLSRNLELIKAVVEDAEEKQ 244
Query: 61 YQIIYVKKWLDKLKHVVYEADQLLDEISTDAM 92
+K WL LK VY D +LDE S +++
Sbjct: 245 ITDKPIKVWLQHLKDAVYVLDDILDECSIESL 340
>BP057104
Length = 496
Score = 44.3 bits (103), Expect = 1e-04
Identities = 22/68 (32%), Positives = 40/68 (58%)
Frame = -3
Query: 566 LVDEISNIKLLRYLDLSFTEITSLPDSICMLYNLQTILLQGCELTELPSNFSKLINLRHL 625
L+ ++ + L++LDLSF + +PD+I L++L+ + L G LP + +L LR L
Sbjct: 341 LLPDLPHFSCLQHLDLSFCNLRQVPDAIGWLHSLEDLDLGGNNFISLPPSIKELSKLRVL 162
Query: 626 ELPYLKKM 633
L + K++
Sbjct: 161 NLEHCKQL 138
Score = 34.3 bits (77), Expect = 0.11
Identities = 27/87 (31%), Positives = 38/87 (43%), Gaps = 4/87 (4%)
Frame = -3
Query: 553 LRTLSFRWCGLSELVDEISNIKLLRYLDLSFTEITSLPDSICMLYNLQTILLQGCE---- 608
L+ L +C L ++ D I + L LDL SLP SI L L+ + L+ C+
Sbjct: 311 LQHLDLSFCNLRQVPDAIGWLHSLEDLDLGGNNFISLPPSIKELSKLRVLNLEHCKQLRY 132
Query: 609 LTELPSNFSKLINLRHLELPYLKKMPK 635
ELPS + R Y+ PK
Sbjct: 131 FPELPSRTQGWVVRRPHAGLYIFNCPK 51
>AV409095
Length = 414
Score = 43.9 bits (102), Expect = 1e-04
Identities = 26/62 (41%), Positives = 35/62 (55%)
Frame = +2
Query: 566 LVDEISNIKLLRYLDLSFTEITSLPDSICMLYNLQTILLQGCELTELPSNFSKLINLRHL 625
L EI +K L YLDLSF +I +LP I L L ++ + +L ELPS + L L L
Sbjct: 11 LPPEIGCLKSLEYLDLSFNKIKTLPTEITYLIGLISMKVANNKLVELPSAMTSLSRLECL 190
Query: 626 EL 627
+L
Sbjct: 191 DL 196
>AV777489
Length = 451
Score = 43.9 bits (102), Expect = 1e-04
Identities = 21/57 (36%), Positives = 34/57 (58%)
Frame = +1
Query: 36 DDLVKELNIALNSINHVLEEAEIKQYQIIYVKKWLDKLKHVVYEADQLLDEISTDAM 92
D+ + L+ L +I LE+AE KQ+ +K WL KL+ + D +LDE +T+A+
Sbjct: 52 DEELTRLSSTLTAIKATLEDAEEKQFTDRAIKVWLQKLRDAAHVLDDILDECATEAL 222
>AV419023
Length = 410
Score = 43.9 bits (102), Expect = 1e-04
Identities = 28/69 (40%), Positives = 36/69 (51%)
Frame = +3
Query: 579 LDLSFTEITSLPDSICMLYNLQTILLQGCELTELPSNFSKLINLRHLELPYLKKMPKHIG 638
LDLSF I +P++I L+ L+ + LQG ELPS S+L L +L L H
Sbjct: 12 LDLSFCSILEVPNAIGRLWCLERLNLQGNNFVELPSTISRLRRLAYLNL-------SHCI 170
Query: 639 KLNSLQTLP 647
KL L LP
Sbjct: 171 KLGDLPLLP 197
>TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodulation aberrant
root formation protein), complete
Length = 3308
Score = 42.4 bits (98), Expect = 4e-04
Identities = 82/350 (23%), Positives = 144/350 (40%), Gaps = 19/350 (5%)
Frame = +2
Query: 547 FSRLNFLRTLSFRWCGLS-ELVDEISNIKLLRYLDLSFTEIT-SLPDSICMLYNLQTILL 604
F + LR L C L+ E+ + N+ L L + +T ++P + + +L ++ L
Sbjct: 827 FGSMENLRLLEMANCNLTGEIPPSLGNLTKLHSLFVQMNNLTGTIPPELSSMMSLMSLDL 1006
Query: 605 QGCELT-ELPSNFSKLINL---RHLELPYLKKMPKHIGKLNSLQTLPY------FVVEEK 654
+LT E+P +FSKL NL + + +P IG L +L+TL FV+
Sbjct: 1007 SINDLTGEIPESFSKLKNLTLMNFFQNKFRGSLPSFIGDLPNLETLQVWENNFSFVLPHN 1186
Query: 655 ---NGSDLKELEKLNHLHGKI----CIDGLGYVFDPEDAVTANLKDKKYLEELYMIFYDR 707
NG L NHL G I C G F +T N + +
Sbjct: 1187 LGGNGRFLYFDVTKNHLTGLIPPDLCKSGRLKTF----IITDNFFRGPIPKGIGECRSLT 1354
Query: 708 KKEVDDSIVESNVSVLEALQPNRSLKRLSISQYRGNRFPNWIRGCHLPNLVSLQMRHCGL 767
K V ++ ++ V P+ ++ LS ++ G P+ I G +L +L + +
Sbjct: 1355 KIRVANNFLDGPVPPGVFQLPSVTITELSNNRLNG-ELPSVISG---ESLGTLTLSNNLF 1522
Query: 768 CSHLPPLGQLPSLRELSISNCKRIKIIGEELYGNNSKIDAFRSLEVLEFQRMENLEEWLC 827
+P + +LR L + + IG E+ G +I + + + +
Sbjct: 1523 TGKIP--AAMKNLRALQSLSLDANEFIG-EIPGGVFEIPMLTKVNISGNNLTGPIPTTIT 1693
Query: 828 HEGFLSLKELTIKDCPKLKRALPQHLPSLQKLSIINCNKLEASMPEGDNI 877
H L+ +L+ + L +P+ + +L LSI+N ++ E S P D I
Sbjct: 1694 HRASLTAVDLSRNN---LAGEVPKGMKNLMDLSILNLSRNEISGPVPDEI 1834
Score = 30.8 bits (68), Expect = 1.2
Identities = 41/149 (27%), Positives = 62/149 (41%), Gaps = 9/149 (6%)
Frame = +2
Query: 722 VLEALQPNRSLKRLSI---SQYRGNRFPNWIRGCHLPNLVSLQMRHCGLCSHLPP-LGQL 777
V E+L ++LK L + + Y G P + + NL L+M +C L +PP LG L
Sbjct: 740 VPESLAKLKTLKELHLGYSNAYEGGIPPAF---GSMENLRLLEMANCNLTGEIPPSLGNL 910
Query: 778 PSLRELSISNCKRIKIIGEELYGNNSKIDAFRSLEVLEFQRMENLEEWLCHEGFLSLKEL 837
L L + I EL S + S+ L + E F LK L
Sbjct: 911 TKLHSLFVQMNNLTGTIPPELSSMMSLMSLDLSINDLTGE---------IPESFSKLKNL 1063
Query: 838 TIKDC--PKLKRALPQ---HLPSLQKLSI 861
T+ + K + +LP LP+L+ L +
Sbjct: 1064TLMNFFQNKFRGSLPSFIGDLPNLETLQV 1150
>TC12566 similar to UP|Q84XI1 (Q84XI1) RCa1 (Fragment), partial (28%)
Length = 918
Score = 41.2 bits (95), Expect = 0.001
Identities = 46/194 (23%), Positives = 82/194 (41%), Gaps = 1/194 (0%)
Frame = +1
Query: 300 DLRQLVKSDCWRLFVTHAFQGKSVCDYPKLESIGRKIMDKCEGLPLAIISLGQLLRKKFS 359
+++++ D +LF +AF+ + + +K++ +G+PLA+ LG L +
Sbjct: 16 EVKEMSFEDSLQLFSLNAFKRNDPLE--TYIDLSKKLLSYAKGIPLALKVLGLFLYGR-E 186
Query: 360 QDEWMKILETDMWRLSDV-DNKINPVLRLSYHNLPSDQKRCFAFCSIFPKGYTFEKDELI 418
+ W E+ + +L + D +I VL+LSY L DQK F + F G+ E
Sbjct: 187 RKAW----ESQLVKLEKLPDPEIINVLKLSYDGLDDDQKDIFLDIACFYLGHLEES---- 342
Query: 419 KLWMAEGLLKCCGSYKSEEEFGNEIFGDLESISFFQQSFDKTYGTYEHYVMYNLVNDLAK 478
L CG + + G E+ D IS + VM++L+ + K
Sbjct: 343 ----VAQTLDSCGFF---ADIGMEVLKDRCLISILE----------GRIVMHDLIEVMGK 471
Query: 479 SVSGEFCMQIEGAR 492
+ + C G R
Sbjct: 472 EIVRQQCPNDPGKR 513
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.321 0.139 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,770,109
Number of Sequences: 28460
Number of extensions: 320577
Number of successful extensions: 1706
Number of sequences better than 10.0: 66
Number of HSP's better than 10.0 without gapping: 1652
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1694
length of query: 1104
length of database: 4,897,600
effective HSP length: 100
effective length of query: 1004
effective length of database: 2,051,600
effective search space: 2059806400
effective search space used: 2059806400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC133863.3


