

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133862.6 - phase: 0
(368 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
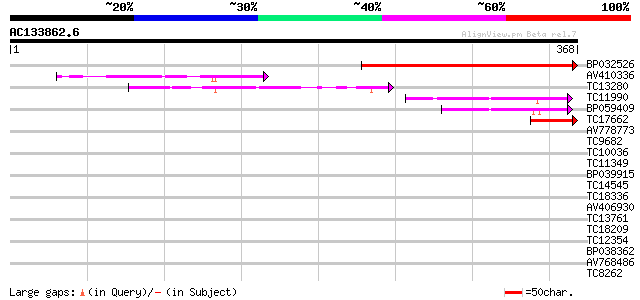
Score E
Sequences producing significant alignments: (bits) Value
BP032526 209 4e-55
AV410336 103 3e-23
TC13280 weakly similar to GB|AAP68292.1|31711872|BT008853 At5g11... 75 1e-14
TC11990 56 1e-08
BP059409 52 2e-07
TC17662 weakly similar to GB|AAP68292.1|31711872|BT008853 At5g11... 44 3e-05
AV778773 35 0.015
TC9682 similar to UP|Q8VZC0 (Q8VZC0) DTDP-glucose 4-6-dehydratas... 32 0.17
TC10036 weakly similar to UP|Q9LFB5 (Q9LFB5) Anthranilate N-benz... 32 0.22
TC11349 similar to UP|Q9SIN1 (Q9SIN1) Expressed protein (At2g425... 31 0.29
BP039915 31 0.29
TC14545 similar to UP|Q9FT78 (Q9FT78) P23 co-chaperone, partial ... 30 0.49
TC18336 similar to UP|Q84KA9 (Q84KA9) RING/C3HC4/PHD zinc finger... 29 1.4
AV406930 29 1.4
TC13761 weakly similar to GB|AAO63290.1|28950733|BT005226 At5g57... 29 1.4
TC18209 similar to UP|TRPD_ARATH (Q02166) Anthranilate phosphori... 29 1.4
TC12354 similar to UP|O23702 (O23702) Dehydrogenase, partial (25%) 28 1.9
BP038362 28 1.9
AV768486 28 1.9
TC8262 homologue to UP|Q8LPE6 (Q8LPE6) Epoxide hydrolase (Fragm... 28 1.9
>BP032526
Length = 516
Score = 209 bits (533), Expect = 4e-55
Identities = 95/140 (67%), Positives = 114/140 (80%)
Frame = -1
Query: 229 ARGRYAMMPEVPFEKFRVGSQFFTLTRKHALVVVKDRTLWRKFKVPCYRDDECYPEEHYF 288
ARG M+PEV F+ FRVGSQFF LTR+HA+VV++DR LWRKF++PC ++ CYPEEHYF
Sbjct: 516 ARGENVMLPEVTFDAFRVGSQFFVLTRRHAVVVLRDRRLWRKFRLPCLTEEPCYPEEHYF 337
Query: 289 PTLLSMEDSDGVTGYTLTNVNWTGTVNGHPHTYQPEEVSPELILRLRKSTNSESFLFARK 348
PTLLSMED +G TG+TLT VNWTG +GHPH Y P EVSPEL+ RLR+S +S S+LFARK
Sbjct: 336 PTLLSMEDPNGCTGFTLTRVNWTGCWDGHPHLYTPPEVSPELVRRLRESNSSYSYLFARK 157
Query: 349 FVPDCLEPLMGIAKSVIFKD 368
F P+CL PLM IA VIF+D
Sbjct: 156 FSPECLTPLMDIADDVIFRD 97
>AV410336
Length = 358
Score = 103 bits (258), Expect = 3e-23
Identities = 67/142 (47%), Positives = 84/142 (58%), Gaps = 4/142 (2%)
Frame = +3
Query: 31 PSPIPISPTDELDDINLFNTAISHSTPPSSSSHPSKFFHLSSKNPTFKIAFLFLTNTDLH 90
PSP T + DD+ LF H + ++ NP KIAFLFLTN++L
Sbjct: 12 PSPF----TSDADDLYLF--------------HRASAARATTTNPKPKIAFLFLTNSNLT 137
Query: 91 FTPLWNLFFQTTPSKLFNVYVHSDPRVNLTLLRSSNNYNP--IF--KFISSKKTYRASPT 146
F PLW FF T L+N+Y+H+DP SS +P +F +FI+SKKTYR+SPT
Sbjct: 138 FAPLWEKFF-TGHHHLYNIYIHADP--------SSAVVSPGGVFHRRFITSKKTYRSSPT 290
Query: 147 LISATRRLLASAILDDASNAYF 168
LISATRRLLASA+LDD N YF
Sbjct: 291 LISATRRLLASAVLDDPLNEYF 356
>TC13280 weakly similar to GB|AAP68292.1|31711872|BT008853 At5g11730
{Arabidopsis thaliana;}, partial (39%)
Length = 702
Score = 75.5 bits (184), Expect = 1e-14
Identities = 58/176 (32%), Positives = 88/176 (49%), Gaps = 4/176 (2%)
Frame = +3
Query: 78 KIAFLFLTNTDLHFTPLWNLFFQTTPSKLFNVYVHSDPRVNLTLLRSSNNYNPIF--KFI 135
K+AFLFL +++ PLW +FF+ F++YVHS P N ++ +P+F + I
Sbjct: 267 KVAFLFLVRSNVPLAPLWEVFFRGHEG-YFSIYVHSHPSYN------GSDKSPLFRGRRI 425
Query: 136 SSKKTYRASPTLISATRRLLASAILDDASNAYFIVLSQYCIPLHSFDYIYKSLFLSPTFD 195
SK ++ A RRLLA+A+L D SN F+++S+ CIPL +F IY L
Sbjct: 426 PSKIVEWGRVNMMEAERRLLANALL-DFSNQRFVLISESCIPLFNFSTIYSYLM------ 584
Query: 196 LTDSESTQFGVRLKYKSFIEIINNGPRLWKRYTARGRY--AMMPEVPFEKFRVGSQ 249
STQ +++ ++ RGRY M PE+ ++R GSQ
Sbjct: 585 ----NSTQ--------NYVMAVDE-----PSAAGRGRYKIQMSPEITLRQWRKGSQ 701
>TC11990
Length = 544
Score = 55.8 bits (133), Expect = 1e-08
Identities = 40/118 (33%), Positives = 58/118 (48%), Gaps = 10/118 (8%)
Frame = +3
Query: 258 ALVVVKDRTLWRKFKVPCYRDDECYPEEHYFPTLLSMEDSDGVTGYTLTNVNWTGTVNGH 317
AL VV DR + F+ C + +CY +EHY PTL++++ + TLT V+WT H
Sbjct: 3 ALQVVSDRRYFPVFEKYC--NPQCYSDEHYLPTLVNIKFWKRNSNRTLTWVDWT-KGGPH 173
Query: 318 PHTYQPEEVSPELILRLRKSTNSE----------SFLFARKFVPDCLEPLMGIAKSVI 365
P Y +V+ E + RLR + LFARKF LE L+ A ++
Sbjct: 174 PSKYIRTDVTVEFLERLRYGNGTTCEYHGMVTNICHLFARKFTTHALERLLRFAPKLM 347
>BP059409
Length = 357
Score = 52.0 bits (123), Expect = 2e-07
Identities = 32/92 (34%), Positives = 51/92 (54%), Gaps = 7/92 (7%)
Frame = -1
Query: 281 CYPEEHYFPTLLSMEDSDGVTGYTLTNVNWTGTVNGHPHTYQPEEVSPELILRLRKST-- 338
CY +EHY PT +++ + + +LT V+W+ HP ++ +V+ E + RLR
Sbjct: 351 CYADEHYLPTFVNIMFGERNSNRSLTWVDWS-RGGPHPASFTRSDVTVEFLERLRSQRCK 175
Query: 339 -NSES----FLFARKFVPDCLEPLMGIAKSVI 365
N +S +LFARKF+P L L+ IA V+
Sbjct: 174 YNGKSSHVCYLFARKFLPSTLSRLVKIAPEVM 79
>TC17662 weakly similar to GB|AAP68292.1|31711872|BT008853 At5g11730
{Arabidopsis thaliana;}, partial (6%)
Length = 513
Score = 44.3 bits (103), Expect = 3e-05
Identities = 20/30 (66%), Positives = 24/30 (79%)
Frame = +1
Query: 339 NSESFLFARKFVPDCLEPLMGIAKSVIFKD 368
+S S+LFARKF P+CL PLM IA VIF+D
Sbjct: 4 SSYSYLFARKFSPECLTPLMDIADDVIFRD 93
>AV778773
Length = 468
Score = 35.4 bits (80), Expect = 0.015
Identities = 16/49 (32%), Positives = 22/49 (44%)
Frame = +1
Query: 15 SLPILFFLAPAILPPHPSPIPISPTDELDDINLFNTAISHSTPPSSSSH 63
++P FF P +LP P P PT + A +HS P + SH
Sbjct: 64 NIPFFFFFTPILLPSTPPRSPFRPTSVAPSPSSTTAAAAHSLPTCTRSH 210
>TC9682 similar to UP|Q8VZC0 (Q8VZC0) DTDP-glucose 4-6-dehydratase-like
protein, partial (29%)
Length = 731
Score = 32.0 bits (71), Expect = 0.17
Identities = 16/36 (44%), Positives = 24/36 (66%)
Frame = +1
Query: 29 PHPSPIPISPTDELDDINLFNTAISHSTPPSSSSHP 64
P PSP P S T ++ ++ +++ S S PPSSSS+P
Sbjct: 286 PDPSPDP-STTSSVNSVSSSSSSESSSAPPSSSSNP 390
>TC10036 weakly similar to UP|Q9LFB5 (Q9LFB5) Anthranilate
N-benzoyltransferase-like protein (AT5g01210/F7J8_190),
partial (32%)
Length = 502
Score = 31.6 bits (70), Expect = 0.22
Identities = 22/59 (37%), Positives = 30/59 (50%), Gaps = 2/59 (3%)
Frame = +3
Query: 8 LTFSLLLSLP--ILFFLAPAILPPHPSPIPISPTDELDDINLFNTAISHSTPPSSSSHP 64
+TF+L + P ILF L P+I P PS P+S T L + T TP ++S P
Sbjct: 285 VTFTLPATTPAWILFTLKPSISPSTPSFRPVSSTCILVSRSFSPTTCRFPTPATTSPSP 461
>TC11349 similar to UP|Q9SIN1 (Q9SIN1) Expressed protein
(At2g42580/F14N22.15), partial (5%)
Length = 474
Score = 31.2 bits (69), Expect = 0.29
Identities = 31/96 (32%), Positives = 43/96 (44%), Gaps = 10/96 (10%)
Frame = +1
Query: 5 PYLLTFSL--------LLSLPILFFLAPAILPPHPSPIPISPTDELDDIN--LFNTAISH 54
P+ LT SL +LSL I F +++ H PIP P L I+ L +
Sbjct: 13 PHSLTLSLSLETKKSFILSLRIHSFFTCSLILYHEIPIPSHPFSSLSTIHSCLVLLQLLL 192
Query: 55 STPPSSSSHPSKFFHLSSKNPTFKIAFLFLTNTDLH 90
ST S+ S+ S F +S +N +AFL T H
Sbjct: 193 STLNSTRSN-SNFLFVSRENQNNNVAFLGKTRFRRH 297
>BP039915
Length = 541
Score = 31.2 bits (69), Expect = 0.29
Identities = 14/25 (56%), Positives = 19/25 (76%), Gaps = 2/25 (8%)
Frame = +3
Query: 11 SLLLSLPILFFLAPAIL--PPHPSP 33
+LLLSLP+LFF P +L P +P+P
Sbjct: 84 TLLLSLPVLFFFVPTLLLDPSNPTP 158
>TC14545 similar to UP|Q9FT78 (Q9FT78) P23 co-chaperone, partial (46%)
Length = 1127
Score = 30.4 bits (67), Expect = 0.49
Identities = 29/94 (30%), Positives = 35/94 (36%), Gaps = 24/94 (25%)
Frame = -1
Query: 53 SHSTPPSSSSHPSKFFHLSSKNPTFKIAF-----------------LFLTNTDLHFTPLW 95
SHST PS S+HPSK S T F L +HF +W
Sbjct: 578 SHSTHPSHSTHPSKSSKTSITAHTHSTKFREVHPSHIRFSNNIFFILIHPFIPIHFHIMW 399
Query: 96 NLFFQT----TPSKLFNVY---VHSDPRVNLTLL 122
L F T PS L ++ S P +N LL
Sbjct: 398 RLAFSTQQPFPPSTLCLLHHREYASHPNINFALL 297
>TC18336 similar to UP|Q84KA9 (Q84KA9) RING/C3HC4/PHD zinc finger-like
protein, partial (15%)
Length = 761
Score = 28.9 bits (63), Expect = 1.4
Identities = 20/70 (28%), Positives = 32/70 (45%)
Frame = -3
Query: 24 PAILPPHPSPIPISPTDELDDINLFNTAISHSTPPSSSSHPSKFFHLSSKNPTFKIAFLF 83
P PH SP+ +D L + IS PP ++ +P + H +S+ P + L
Sbjct: 396 PGFPLPHSSPLHQPKSDPL-------SVISQKHPPLTTHYPPQHTHQNSRTPHHG-SHLP 241
Query: 84 LTNTDLHFTP 93
L + + H TP
Sbjct: 240 LLSQNFHQTP 211
>AV406930
Length = 433
Score = 28.9 bits (63), Expect = 1.4
Identities = 24/68 (35%), Positives = 32/68 (46%), Gaps = 5/68 (7%)
Frame = +1
Query: 14 LSLPILFFLAPAILPPHPSPIPISPTDELDD--INLFNTAISHS---TPPSSSSHPSKFF 68
L+L FLAP P P+P IS + I F+T +SHS P +S P F
Sbjct: 58 LALAPPHFLAPPPPPQPPNPPSISQNLSISTHPITPFHTPLSHSFNLPLPKPASIPLSFS 237
Query: 69 HLSSKNPT 76
L S +P+
Sbjct: 238 ALPSPSPS 261
>TC13761 weakly similar to GB|AAO63290.1|28950733|BT005226 At5g57910
{Arabidopsis thaliana;}, partial (21%)
Length = 652
Score = 28.9 bits (63), Expect = 1.4
Identities = 24/63 (38%), Positives = 29/63 (45%)
Frame = +1
Query: 20 FFLAPAILPPHPSPIPISPTDELDDINLFNTAISHSTPPSSSSHPSKFFHLSSKNPTFKI 79
FFL P+ + HP + LF + + STPPSSSS FF LS TF
Sbjct: 115 FFLTPSFIHTHPF-----------SLFLFLSHVFSSTPPSSSS----FFFLSICFFTFCS 249
Query: 80 AFL 82
FL
Sbjct: 250 WFL 258
>TC18209 similar to UP|TRPD_ARATH (Q02166) Anthranilate
phosphoribosyltransferase, chloroplast precursor ,
partial (41%)
Length = 774
Score = 28.9 bits (63), Expect = 1.4
Identities = 21/63 (33%), Positives = 30/63 (47%)
Frame = +3
Query: 97 LFFQTTPSKLFNVYVHSDPRVNLTLLRSSNNYNPIFKFISSKKTYRASPTLISATRRLLA 156
LF T S F H P NLTLLR+ +PI +S+ + A + I + +L+
Sbjct: 48 LFSTLTSSPSFFFTHHRVPSPNLTLLRTPPLPSPISHRVSAAVSCNAGVSSIKSVPQLIE 227
Query: 157 SAI 159
S I
Sbjct: 228 SLI 236
>TC12354 similar to UP|O23702 (O23702) Dehydrogenase, partial (25%)
Length = 488
Score = 28.5 bits (62), Expect = 1.9
Identities = 17/47 (36%), Positives = 21/47 (44%)
Frame = +3
Query: 23 APAILPPHPSPIPISPTDELDDINLFNTAISHSTPPSSSSHPSKFFH 69
A I P +P P SP ++ + S PP SSS PS FH
Sbjct: 39 ASKIAPSSSNPSPASPPSSTS-LSAVSPMARSSPPPLSSSTPSPTFH 176
>BP038362
Length = 274
Score = 28.5 bits (62), Expect = 1.9
Identities = 19/46 (41%), Positives = 21/46 (45%)
Frame = -3
Query: 26 ILPPHPSPIPISPTDELDDINLFNTAISHSTPPSSSSHPSKFFHLS 71
IL P SP P SPT E + H T P SSHPS L+
Sbjct: 230 ILDPQ-SPFPFSPTVEAFSY------LGHGTSPWESSHPSPTLDLT 114
>AV768486
Length = 398
Score = 28.5 bits (62), Expect = 1.9
Identities = 22/77 (28%), Positives = 33/77 (42%)
Frame = +3
Query: 15 SLPILFFLAPAILPPHPSPIPISPTDELDDINLFNTAISHSTPPSSSSHPSKFFHLSSKN 74
S P + LAP+ LPP P+ PT E +IS +P S P F S++
Sbjct: 135 SSPNFYLLAPSPLPPSSPPVQ-HPTKE--------GSISFQSPHSLIFDPCVIFPSHSEH 287
Query: 75 PTFKIAFLFLTNTDLHF 91
P + + + +HF
Sbjct: 288 PPTFMPWNVCIHISIHF 338
>TC8262 homologue to UP|Q8LPE6 (Q8LPE6) Epoxide hydrolase (Fragment) ,
partial (13%)
Length = 379
Score = 28.5 bits (62), Expect = 1.9
Identities = 15/50 (30%), Positives = 27/50 (54%)
Frame = -2
Query: 78 KIAFLFLTNTDLHFTPLWNLFFQTTPSKLFNVYVHSDPRVNLTLLRSSNN 127
+++ FL N HF +W++FF+ S + N+ +H + V+ L S N
Sbjct: 150 EVSHSFLHN---HFLQIWHIFFE---SSIMNILLHVERGVDQXLASSDMN 19
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.323 0.138 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,710,794
Number of Sequences: 28460
Number of extensions: 133466
Number of successful extensions: 1210
Number of sequences better than 10.0: 71
Number of HSP's better than 10.0 without gapping: 1175
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1202
length of query: 368
length of database: 4,897,600
effective HSP length: 92
effective length of query: 276
effective length of database: 2,279,280
effective search space: 629081280
effective search space used: 629081280
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 56 (26.2 bits)
Medicago: description of AC133862.6


