

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133780.9 - phase: 0 /pseudo
(562 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
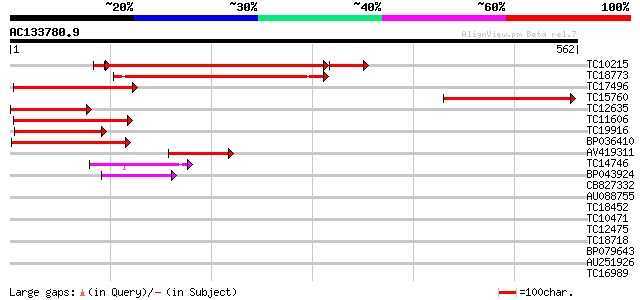
Score E
Sequences producing significant alignments: (bits) Value
TC10215 similar to UP|Q9LW29 (Q9LW29) Transport inhibitor respon... 377 e-113
TC18773 similar to PIR|T48087|T48087 transport inhibitor respons... 231 3e-61
TC17496 similar to PIR|T48087|T48087 transport inhibitor respons... 175 2e-44
TC15760 similar to UP|Q9LW29 (Q9LW29) Transport inhibitor respon... 163 8e-41
TC12635 similar to UP|Q9LW29 (Q9LW29) Transport inhibitor respon... 157 3e-39
TC11606 weakly similar to UP|Q9LW29 (Q9LW29) Transport inhibitor... 150 7e-37
TC19916 similar to PIR|T48087|T48087 transport inhibitor respons... 143 8e-35
BP036410 107 4e-24
AV419311 89 1e-18
TC14746 weakly similar to GB|AAG21976.1|10716947|AF263377 SKP1 i... 42 3e-04
BP043924 41 4e-04
CB827332 39 0.002
AU088755 36 0.019
TC18452 weakly similar to GB|AAO64875.1|29028992|BT005940 At5g27... 35 0.025
TC10471 weakly similar to UP|Q84QE1 (Q84QE1) Avr9/Cf-9 rapidly e... 33 0.12
TC12475 weakly similar to UP|O22512 (O22512) Grr1, partial (23%) 32 0.27
TC18718 28 3.9
BP079643 28 5.1
AU251926 28 5.1
TC16989 similar to PIR|S66350|S66350 lycopene beta-cyclase - to... 28 5.1
>TC10215 similar to UP|Q9LW29 (Q9LW29) Transport inhibitor response-like
protein, partial (43%)
Length = 837
Score = 377 bits (969), Expect(3) = e-113
Identities = 193/222 (86%), Positives = 205/222 (91%)
Frame = +3
Query: 95 LAKNKVGLEELRLKRMVVSDESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLR 154
LAKNKVGLEELRLKRMVVSDESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCR LR
Sbjct: 33 LAKNKVGLEELRLKRMVVSDESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRFLR 212
Query: 155 ELDLQENEVEDHKGQWLSCFPESCTSLVSLNFACLKGDINLGALERLVSRSPNLKSLRLN 214
ELDLQENEVED KGQWLSCFP+SCTSLVSLNFACLKGD+NLGALERLV+RSPNLKSLRLN
Sbjct: 213 ELDLQENEVEDQKGQWLSCFPDSCTSLVSLNFACLKGDVNLGALERLVARSPNLKSLRLN 392
Query: 215 RSVPVDALQRILTRAPQLMDLGIGSFFHDLNSDAYAMFKATILKCKSITSLSGFLEVAPF 274
RSVP++ALQRIL +APQL DLGIGSF H+ SDA++ K TILK KSITSLSGFLEV PF
Sbjct: 393 RSVPIEALQRILMQAPQLADLGIGSFVHNPFSDAFSKLKNTILKSKSITSLSGFLEVGPF 572
Query: 275 SLAAIYPICQNLTSLNLSYAAGILGIELIKLIRHCGKLQRLW 316
L A+YPIC+NLT+LNLSYAAGI G ELIKLI HCGKLQRLW
Sbjct: 573 CLPAMYPICRNLTALNLSYAAGIHGNELIKLIYHCGKLQRLW 698
Score = 39.3 bits (90), Expect(3) = e-113
Identities = 23/38 (60%), Positives = 24/38 (62%)
Frame = +1
Query: 318 D*VL*LAHVKNCKN*EFSHQLHLETKQLLPKKDL*RYQ 355
D VL L HVKNCKN* SH L LE L KKD +YQ
Sbjct: 724 DWVLWLVHVKNCKN*GSSHLLPLEILHLSLKKDWLQYQ 837
Score = 32.7 bits (73), Expect(3) = e-113
Identities = 11/17 (64%), Positives = 14/17 (81%)
Frame = +1
Query: 84 WGGFVYPWIEALAKNKV 100
WGGFVYPWIEA + ++
Sbjct: 1 WGGFVYPWIEAWPRTRL 51
Score = 26.9 bits (58), Expect = 8.8
Identities = 51/182 (28%), Positives = 67/182 (36%), Gaps = 17/182 (9%)
Frame = +3
Query: 54 ERLVERFPDLKSLTLKGK-------------PHFADFSLVPHGWGGFVY-PWIEALAKNK 99
ERLV R P+LKSL L P AD G G FV+ P+ +A +K
Sbjct: 345 ERLVARSPNLKSLRLNRSVPIEALQRILMQAPQLADL-----GIGSFVHNPFSDAFSK-- 503
Query: 100 VGLEELRLKRMVVSDESLELLSRSFVNFKSLVLVSCEGFTTDG---LAAVAANCRSLREL 156
LK ++ +S+ LS GF G L A+ CR+L L
Sbjct: 504 -------LKNTILKSKSITSLS---------------GFLEVGPFCLPAMYPICRNLTAL 617
Query: 157 DLQENEVEDHKGQWLSCFPESCTSLVSLNFACLKGDINLGALERLVSRSPNLKSLRLNRS 216
+L + G L C L L GD LG + S L+ LR+ S
Sbjct: 618 NL--SYAAGIHGNELIKLIYHCGKLQRLWIMDCIGDKGLGI---VASTCKELQELRVFPS 782
Query: 217 VP 218
P
Sbjct: 783 AP 788
>TC18773 similar to PIR|T48087|T48087 transport inhibitor response protein
TIR1 [imported] - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (50%)
Length = 927
Score = 231 bits (588), Expect = 3e-61
Identities = 118/213 (55%), Positives = 154/213 (71%)
Frame = +3
Query: 104 ELRLKRMVVSDESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEV 163
E+RL+ + D+ L+L+++SF NF+ LVL+SCEGFTT GLAA+AANCR+LRELDL+E+EV
Sbjct: 15 EIRLRGWLY-DDXLDLIAKSFKNFRVLVLISCEGFTTHGLAAIAANCRNLRELDLRESEV 191
Query: 164 EDHKGQWLSCFPESCTSLVSLNFACLKGDINLGALERLVSRSPNLKSLRLNRSVPVDALQ 223
ED G WLS FP+S SL SLN +CL ++NL ALERLVSR PNL++LRLNR+VP+D L
Sbjct: 192 EDICGHWLSHFPDSYNSLESLNISCLSNEVNLPALERLVSRCPNLQTLRLNRAVPLDRLT 371
Query: 224 RILTRAPQLMDLGIGSFFHDLNSDAYAMFKATILKCKSITSLSGFLEVAPFSLAAIYPIC 283
+L APQL++LG G++ ++ + A CK + LSGF +V P L A+YP+C
Sbjct: 372 NLLRGAPQLVELGTGAYTAEMRPEVLANLTEAFSGCKQLKGLSGFWDVLPSYLPAVYPVC 551
Query: 284 QNLTSLNLSYAAGILGIELIKLIRHCGKLQRLW 316
LTSLNLSYA I + IKL+ CG LQRLW
Sbjct: 552 SGLTSLNLSYAT-IQSPDHIKLVSQCGSLQRLW 647
Score = 30.4 bits (67), Expect = 0.79
Identities = 24/80 (30%), Positives = 36/80 (45%), Gaps = 8/80 (10%)
Frame = +3
Query: 142 GLAAVAANCRSLREL--------DLQENEVEDHKGQWLSCFPESCTSLVSLNFACLKGDI 193
GL +AA+C+ LREL L+ N +G L E C L S+ + C + +
Sbjct: 672 GLDVLAASCKDLRELRVFPSDPFGLEPNVALTEEG--LISVSEGCPKLQSVLYFCRQ--M 839
Query: 194 NLGALERLVSRSPNLKSLRL 213
+ AL + PN+ RL
Sbjct: 840 SNAALNTIAQNRPNMTRFRL 899
>TC17496 similar to PIR|T48087|T48087 transport inhibitor response protein
TIR1 [imported] - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (22%)
Length = 535
Score = 175 bits (444), Expect = 2e-44
Identities = 75/123 (60%), Positives = 105/123 (84%)
Frame = +3
Query: 4 FPDEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERFPDL 63
FP+EV+EHVF ++ +DRN++SLVCKSWY IER+ R++VF+GNCY++SP +++RFP++
Sbjct: 165 FPEEVLEHVFSFIQVDTDRNAISLVCKSWYEIERWCRRKVFVGNCYAVSPMIVIKRFPEV 344
Query: 64 KSLTLKGKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMVVSDESLELLSRS 123
+S+ LKGKPHFADF+LVP GWGG+V WI A+++ LEE+RLKRMV+SDESLEL+++S
Sbjct: 345 RSIALKGKPHFADFNLVPEGWGGYVCTWIAAMSRAFPWLEEIRLKRMVISDESLELIAKS 524
Query: 124 FVN 126
F N
Sbjct: 525 FKN 533
>TC15760 similar to UP|Q9LW29 (Q9LW29) Transport inhibitor response-like
protein, partial (17%)
Length = 664
Score = 163 bits (412), Expect = 8e-41
Identities = 81/131 (61%), Positives = 97/131 (73%)
Frame = +3
Query: 431 PTRFSFTLVCMLSSLKCYLLHLLARVTRECSMY*TDAKRFASLRLETVLSETRRF*QT*G 490
PTR S TL C LSSL+C L L RVTRECSM *TDA+ F + RL + LS T F +T*G
Sbjct: 3 PTRCSSTLACTLSSLRCCPLRLPGRVTRECSMC*TDARSFGNSRLGSALSATWHFWRT*G 182
Query: 491 SMKQCDPFGCRRAR*L*KHARHWQRRCRG*MWRFSVKVNRQIVTWKMGRESRRCICIVQW 550
SMKQCDPFGC +*L +HAR+ +RRCRG*MWR S++++++ V WKM R RRCIC V W
Sbjct: 183 SMKQCDPFGCHPVK*LWEHARNSRRRCRG*MWRSSMRMSKKSVVWKMSRV*RRCICTVHW 362
Query: 551 LGKGRMHQTMY 561
LG+G MHQ MY
Sbjct: 363 LGRGMMHQNMY 395
>TC12635 similar to UP|Q9LW29 (Q9LW29) Transport inhibitor response-like
protein, partial (14%)
Length = 696
Score = 157 bits (398), Expect = 3e-39
Identities = 72/81 (88%), Positives = 78/81 (95%)
Frame = -1
Query: 1 MNYFPDEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERF 60
MNYFPDEVIEHVFDYVVSH DRN+LSLVCKSWYRIE+ TR+RVFIGNCYSISPERL++RF
Sbjct: 246 MNYFPDEVIEHVFDYVVSHRDRNALSLVCKSWYRIEKSTRKRVFIGNCYSISPERLIQRF 67
Query: 61 PDLKSLTLKGKPHFADFSLVP 81
P L+SLTLKGKPHFADFSLVP
Sbjct: 66 PSLRSLTLKGKPHFADFSLVP 4
>TC11606 weakly similar to UP|Q9LW29 (Q9LW29) Transport inhibitor
response-like protein, partial (18%)
Length = 892
Score = 150 bits (378), Expect = 7e-37
Identities = 71/118 (60%), Positives = 87/118 (73%)
Frame = +2
Query: 4 FPDEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERFPDL 63
FPDEV+E V V S DR+S+SLVCK WY ER++R+ VFIGNCYS+SPE L RFP++
Sbjct: 536 FPDEVLERVLGMVKSRKDRSSVSLVCKEWYNAERWSRRSVFIGNCYSVSPEILTRRFPNI 715
Query: 64 KSLTLKGKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMVVSDESLELLS 121
+S+TLKGKP F+DF+LVP WG + W+ A LEELRLKRM VSDESLE L+
Sbjct: 716 RSVTLKGKPRFSDFNLVPANWGADIRSWLVVFADKYPLLEELRLKRMTVSDESLEFLA 889
>TC19916 similar to PIR|T48087|T48087 transport inhibitor response protein
TIR1 [imported] - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (16%)
Length = 446
Score = 143 bits (360), Expect = 8e-35
Identities = 59/92 (64%), Positives = 80/92 (86%)
Frame = +1
Query: 5 PDEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERFPDLK 64
P+EV+EHVF ++ + DR+S+SLVC+SWY IER R+ VF+GNCY++SPE +V+RFP ++
Sbjct: 142 PEEVLEHVFSFIDNDKDRSSISLVCRSWYEIERCCRRNVFVGNCYAVSPEMVVKRFPRVR 321
Query: 65 SLTLKGKPHFADFSLVPHGWGGFVYPWIEALA 96
S+TLKGKPHFADF+LVP GWGG+V PWI+A+A
Sbjct: 322 SVTLKGKPHFADFNLVPDGWGGYVSPWIKAMA 417
>BP036410
Length = 550
Score = 107 bits (268), Expect = 4e-24
Identities = 52/118 (44%), Positives = 72/118 (60%)
Frame = +3
Query: 2 NYFPDEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSISPERLVERFP 61
N DEV++ V Y+ DR+++S VCK WY ++ TR+ V I CY+ +P RL RFP
Sbjct: 195 NRLVDEVLDCVIPYIDDPKDRDAVSQVCKCWYELDSLTRKHVTIALCYTTTPARLRRRFP 374
Query: 62 DLKSLTLKGKPHFADFSLVPHGWGGFVYPWIEALAKNKVGLEELRLKRMVVSDESLEL 119
L+SL LKGKP A F+L+P WGG V PW+ + + L+ L +RM+V D L L
Sbjct: 375 HLESLKLKGKPRAAMFNLIPEDWGGHVTPWVLEINQYFDCLKSLHFRRMIVKDNDLLL 548
>AV419311
Length = 203
Score = 89.4 bits (220), Expect = 1e-18
Identities = 42/65 (64%), Positives = 53/65 (80%)
Frame = +2
Query: 158 LQENEVEDHKGQWLSCFPESCTSLVSLNFACLKGDINLGALERLVSRSPNLKSLRLNRSV 217
LQE+EVED G WLS FP+S TSLVSLN +CL +++L ALERL+ R PN+K+LRLNR+V
Sbjct: 2 LQESEVEDLSGHWLSHFPDSYTSLVSLNISCLSNEVSLSALERLLGRCPNMKTLRLNRAV 181
Query: 218 PVDAL 222
P+D L
Sbjct: 182 PLDRL 196
>TC14746 weakly similar to GB|AAG21976.1|10716947|AF263377 SKP1 interacting
partner 1 {Arabidopsis thaliana;}, partial (62%)
Length = 719
Score = 42.0 bits (97), Expect = 3e-04
Identities = 29/105 (27%), Positives = 50/105 (47%), Gaps = 3/105 (2%)
Frame = +3
Query: 80 VPHGWGGFVYPWIEALAKNKVGLEELRLKRMVV---SDESLELLSRSFVNFKSLVLVSCE 136
+P W I+++ ++ V + L ++ + SD SL L++ N + L + SC
Sbjct: 273 LPRWWSPEFEAKIDSMLRSVVQWTHIFLTQIRIRHCSDRSLALVAERCPNLEVLSIRSCP 452
Query: 137 GFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCFPESCTSL 181
T D ++ +A NC LRELD+ HK L+ +C +L
Sbjct: 453 HVTDDSISRIAVNCPKLRELDISYCYDVTHKS--LALIGRNCPNL 581
>BP043924
Length = 546
Score = 41.2 bits (95), Expect = 4e-04
Identities = 30/77 (38%), Positives = 43/77 (54%), Gaps = 3/77 (3%)
Frame = +3
Query: 92 IEALAKNKVGLEELRLKRMV-VSDESLELLSRSF-VNFKSLVLVSCEGFTTDGLAAVAAN 149
+ A+A +EEL L R V DESL L++ S+ + + L FT +GL +VA +
Sbjct: 243 LPAMAARYPSVEELDLSRCPRVGDESLTLVAASYRETLRRVDLSRSRFFTGNGLLSVAVS 422
Query: 150 CRSLRELDLQE-NEVED 165
CR+L ELDL E+ D
Sbjct: 423 CRNLVELDLSNATEIRD 473
>CB827332
Length = 511
Score = 39.3 bits (90), Expect = 0.002
Identities = 26/105 (24%), Positives = 57/105 (53%), Gaps = 2/105 (1%)
Frame = +3
Query: 112 VSDESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWL 171
+ DE++ L++ N ++L++ C + D + ++A CR+ + +L+ + + L
Sbjct: 12 IGDETILSLAKFCDNLETLIIGRCRDVSDDAIKSLATACRTNLK-NLRMDWCLNISDSSL 188
Query: 172 SCFPESCTSLVSLNFACLKGDINLGALERLVSRSP--NLKSLRLN 214
SC C +L +L+ AC + +++ A + + S P NLK L+++
Sbjct: 189 SCILSQCRNLEALDIACCE-EVSDAAFQNIGSGEPGLNLKILKVS 320
>AU088755
Length = 431
Score = 35.8 bits (81), Expect = 0.019
Identities = 15/46 (32%), Positives = 26/46 (55%)
Frame = +2
Query: 1 MNYFPDEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIG 46
M+ P+ ++ + + SDRNS+SLVCK Y ++ R + +G
Sbjct: 41 MDNIPEHLVWEILSRIKKTSDRNSVSLVCKRLYNLDNGQRNSLKVG 178
>TC18452 weakly similar to GB|AAO64875.1|29028992|BT005940 At5g27920
{Arabidopsis thaliana;}, partial (11%)
Length = 698
Score = 35.4 bits (80), Expect = 0.025
Identities = 46/192 (23%), Positives = 81/192 (41%), Gaps = 5/192 (2%)
Frame = +2
Query: 129 SLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCFPESCTSLVSLNFA- 187
SL + SC + + C L ELD+ +NEV+D + +S +C+ L SL
Sbjct: 2 SLKMESCTLVPQEAFVLIGQKCHYLEELDITDNEVDDEGLKSIS----NCSRLSSLKVGI 169
Query: 188 CLKGDINLGALERLVSRSPNLKSLRLNRSVPVD--ALQRILTRAPQLMDLGIGSFFHDLN 245
CL +I + + LK L L RS + + I P L + S+ ++
Sbjct: 170 CL--NITDRGVAYIGMCCSKLKELDLYRSTGITDLGISAIACGCPGLEIINT-SYCTNIT 340
Query: 246 SDAYAMFKATILKCKSITSLS--GFLEVAPFSLAAIYPICQNLTSLNLSYAAGILGIELI 303
+ ++ KC+++ +L G L V LA+I C+ L +++ I +I
Sbjct: 341 DGSL----ISLSKCRNLKTLEIRGCLLVTSIGLASIAMNCKQLICVDIKKCYNIDDSGMI 508
Query: 304 KLIRHCGKLQRL 315
L L+++
Sbjct: 509 PLAYFSKSLRQI 544
Score = 32.3 bits (72), Expect = 0.21
Identities = 24/70 (34%), Positives = 39/70 (55%), Gaps = 1/70 (1%)
Frame = +2
Query: 92 IEALAKNKVGLEELRLKRMV-VSDESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANC 150
I A+A GLE + ++D SL LS+ N K+L + C T+ GLA++A NC
Sbjct: 272 ISAIACGCPGLEIINTSYCTNITDGSLISLSKCR-NLKTLEIRGCLLVTSIGLASIAMNC 448
Query: 151 RSLRELDLQE 160
+ L +D+++
Sbjct: 449 KQLICVDIKK 478
>TC10471 weakly similar to UP|Q84QE1 (Q84QE1) Avr9/Cf-9 rapidly elicited
protein 189, partial (18%)
Length = 459
Score = 33.1 bits (74), Expect = 0.12
Identities = 19/67 (28%), Positives = 37/67 (54%), Gaps = 2/67 (2%)
Frame = +1
Query: 5 PDEVIEHVFDYVVSHSDRNSLSLVCKSWYRIERFTRQRVFIGNCYSI--SPERLVERFPD 62
PDE + +F ++++ +DR S VC+ W R++ R+R+ + S+ + L RF
Sbjct: 184 PDECLAAIF-HLLTTTDRKICSAVCRRWLRVDGENRRRLSLNAEASLLDAIPSLFSRFDS 360
Query: 63 LKSLTLK 69
+ L+L+
Sbjct: 361 VTKLSLR 381
>TC12475 weakly similar to UP|O22512 (O22512) Grr1, partial (23%)
Length = 501
Score = 32.0 bits (71), Expect = 0.27
Identities = 19/57 (33%), Positives = 29/57 (50%)
Frame = +2
Query: 128 KSLVLVSCEGFTTDGLAAVAANCRSLRELDLQENEVEDHKGQWLSCFPESCTSLVSL 184
KSL + SC G T G+ AV C +L+ + L + G L F ++ +SL +L
Sbjct: 98 KSLTIASCRGVTDVGIEAVGKGCPNLKSVHLHKCAFLSDNG--LISFTKAASSLENL 262
>TC18718
Length = 518
Score = 28.1 bits (61), Expect = 3.9
Identities = 14/37 (37%), Positives = 22/37 (58%), Gaps = 1/37 (2%)
Frame = +1
Query: 280 YPICQNLTSLNLSYAAGILG-IELIKLIRHCGKLQRL 315
+P NLT + L Y I + +++L++HC KLQ L
Sbjct: 103 FPKFHNLTHIELEYNCRISDWLTVVELLKHCPKLQVL 213
>BP079643
Length = 451
Score = 27.7 bits (60), Expect = 5.1
Identities = 21/82 (25%), Positives = 43/82 (51%), Gaps = 3/82 (3%)
Frame = -3
Query: 83 GWGGFVYPWIEALAKNKVGLEELR--LKRMVVSDESLELLSRSF-VNFKSLVLVSCEGFT 139
G G + P +EA + +G +E+ ++ ++V +E ++ R+ + +L V C G +
Sbjct: 359 GLGVAIKPVVEA-GRKVIGRKEIERVVREVMVGEEGKKMRERARELKESALKTVQCGGKS 183
Query: 140 TDGLAAVAANCRSLRELDLQEN 161
+ LA V + R + L ++EN
Sbjct: 182 YEALANVVGHVRMIN*LMVEEN 117
>AU251926
Length = 335
Score = 27.7 bits (60), Expect = 5.1
Identities = 22/94 (23%), Positives = 42/94 (44%)
Frame = +2
Query: 99 KVGLEELRLKRMVVSDESLELLSRSFVNFKSLVLVSCEGFTTDGLAAVAANCRSLRELDL 158
K G +L+L+ ++ + ++ L S SLV + L + SL LDL
Sbjct: 50 KKGTRDLKLQNKLL--DQVDWLPDSLGKLSSLVTLDLSENRIVALPSTIGGLSSLTRLDL 223
Query: 159 QENEVEDHKGQWLSCFPESCTSLVSLNFACLKGD 192
N +++ P+S +L++L + L+G+
Sbjct: 224 HTNRIQE--------LPDSIGNLLNLVYLDLRGN 301
>TC16989 similar to PIR|S66350|S66350 lycopene beta-cyclase - tomato
{Lycopersicon esculentum;} , partial (31%)
Length = 591
Score = 27.7 bits (60), Expect = 5.1
Identities = 25/65 (38%), Positives = 29/65 (44%), Gaps = 2/65 (3%)
Frame = +1
Query: 26 SLVCKSWYRIERFTRQRVFIGNCYSISPERLVERFPDLKSLTLKGKPHFAD--FSLVPHG 83
SLV K + IER RQR F C+ + L L L G F D F L PH
Sbjct: 160 SLVWKDLWPIER-RRQREFF--CFGMDI---------LLKLDLPGTRRFFDAFFDLEPHY 303
Query: 84 WGGFV 88
W GF+
Sbjct: 304 WHGFL 318
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.340 0.146 0.490
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,153,121
Number of Sequences: 28460
Number of extensions: 168639
Number of successful extensions: 1317
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 1300
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1315
length of query: 562
length of database: 4,897,600
effective HSP length: 95
effective length of query: 467
effective length of database: 2,193,900
effective search space: 1024551300
effective search space used: 1024551300
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC133780.9


