

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133780.4 - phase: 1
(377 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
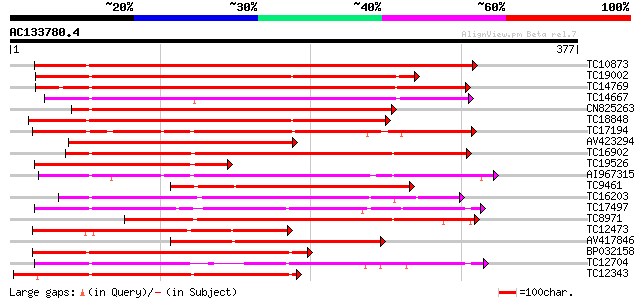
Score E
Sequences producing significant alignments: (bits) Value
TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partia... 388 e-109
TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866)... 233 3e-62
TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete 231 1e-61
TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, com... 229 4e-61
CN825263 228 1e-60
TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, pa... 228 1e-60
TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete 223 3e-59
AV423294 219 4e-58
TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-... 215 1e-56
TC19526 homologue to UP|AAQ96340 (AAQ96340) Protein kinase-like ... 208 1e-54
AI967315 203 3e-53
TC9461 similar to UP|O49840 (O49840) Protein kinase (AT2G02800/T... 193 3e-50
TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodula... 188 1e-48
TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2,... 185 1e-47
TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P... 174 3e-44
TC12473 similar to UP|Q9M068 (Q9M068) Protein kinase-like protei... 170 4e-43
AV417846 169 5e-43
BP032158 167 3e-42
TC12704 UP|CAE45596 (CAE45596) S-receptor kinase-like protein 3,... 166 8e-42
TC12343 similar to UP|CDK9_SCHPO (Q96WV9) Serine/threonine prote... 159 9e-40
>TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partial (77%)
Length = 1478
Score = 388 bits (997), Expect = e-109
Identities = 193/296 (65%), Positives = 236/296 (79%), Gaps = 1/296 (0%)
Frame = +1
Query: 17 AQIFTFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGFQGEKEFLV 76
A F FRELA AT+NF++ IG+GGFG VYKG+L +TG+AVAVK+L G QG +EF++
Sbjct: 406 AASFGFRELADATRNFKEANLIGEGGFGKVYKGRL-TTGEAVAVKQLSHDGRQGFQEFVM 582
Query: 77 EVLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLDWNTRMRI 136
EVLMLSLLHH NLV +IGYC +GDQRLLVYEYMPMGSLE HL +L D EPL+W+TRM++
Sbjct: 583 EVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKV 762
Query: 137 AVGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQSYVATRV 196
AVGAARGL YLH A+P VIYRDLKS+NILLD F PKLSDFGLAK GP GD ++V+TRV
Sbjct: 763 AVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRV 942
Query: 197 MGTHGYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYDET-RAHDKHLVDWARPLFR 255
MGT+GYCAPEYA +GKLT++SDIYSFGVVLLEL+TGRRA D + R +++LV WARP F
Sbjct: 943 MGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPYFS 1122
Query: 256 DKGNFRKLVDPHLQGHYPISGLRMALEMARMCLREDPRLRPSAGDIVLALDYLSSK 311
D+ F +VDP LQG +P L A+ + MCL+E P+ RP DIV+AL+YL+S+
Sbjct: 1123DRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALEYLASQ 1290
>TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866), partial
(48%)
Length = 780
Score = 233 bits (595), Expect = 3e-62
Identities = 122/256 (47%), Positives = 169/256 (65%), Gaps = 1/256 (0%)
Frame = +1
Query: 18 QIFTFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGFQGEKEFLVE 77
++F+ +EL +AT NF + +G+GGFG+VY G+L G +AVKRL + + EF VE
Sbjct: 22 RVFSLKELHSATNNFNYDNKLGEGGFGSVYWGQLWD-GSQIAVKRLKVWSNKADMEFAVE 198
Query: 78 VLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLDWNTRMRIA 137
V +L+ + H NL+S+ GYCAEG +RL+VY+YMP SL SHLH LDWN RM IA
Sbjct: 199 VEILARVRHKNLLSLRGYCAEGQERLIVYDYMPNLSLLSHLHGQHSSECLLDWNRRMNIA 378
Query: 138 VGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQSYVATRVM 197
+G+A G+ YLHH+A P +I+RD+K+SN+LLD F +++DFG AK P G ++V TRV
Sbjct: 379 IGSAEGIVYLHHQATPHIIHRDIKASNVLLDSDFQARVADFGFAKLIPDG-ATHVTTRVK 555
Query: 198 GTHGYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYDETRAHDKHLV-DWARPLFRD 256
GT GY APEYA GK D++SFG++LLEL +G++ ++ + K + DWA PL
Sbjct: 556 GTLGYLAPEYAMLGKANECCDVFSFGILLLELASGKKPLEKLSSTVKRSINDWALPLACA 735
Query: 257 KGNFRKLVDPHLQGHY 272
K F + DP L G Y
Sbjct: 736 K-KFTEFADPRLNGEY 780
>TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete
Length = 3495
Score = 231 bits (590), Expect = 1e-61
Identities = 130/290 (44%), Positives = 178/290 (60%), Gaps = 1/290 (0%)
Frame = +3
Query: 18 QIFTFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGFQGEKEFLVE 77
Q FT + AT+ ++ T IG+GGFG+VY+G L GQ VAVK +T QG +EF E
Sbjct: 2127 QAFTLEYIEVATERYK--TLIGEGGFGSVYRGTLND-GQEVAVKVRSSTSTQGTREFDNE 2297
Query: 78 VLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLDWNTRMRIA 137
+ +LS + H NLV ++GYC E DQ++LVY +M GSL+ L+ + LDW TR+ IA
Sbjct: 2298 LNLLSAIQHENLVPLLGYCNESDQQILVYPFMSNGSLQDRLYGEPAKRKILDWPTRLSIA 2477
Query: 138 VGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQSYVATRVM 197
+GAARGL YLH SVI+RD+KSSNILLD K++DFG +K+ P SYV+ V
Sbjct: 2478 LGAARGLAYLHTFPGRSVIHRDIKSSNILLDHSMCAKVADFGFSKYAPQEGDSYVSLEVR 2657
Query: 198 GTHGYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYDETRAHDK-HLVDWARPLFRD 256
GT GY PEY T +L+ +SD++SFGVVLLE+++GR + R + LV+WA P R
Sbjct: 2658 GTAGYLDPEYYKTQQLSEKSDVFSFGVVLLEIVSGREPLNIKRPRTEWSLVEWATPYIRG 2837
Query: 257 KGNFRKLVDPHLQGHYPISGLRMALEMARMCLREDPRLRPSAGDIVLALD 306
++VDP ++G Y + +E+A CL RPS IV L+
Sbjct: 2838 -SKVDEIVDPGIKGGYHAEAMWRVVEVALQCLEPFSTYRPSMVAIVRELE 2984
>TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, complete
Length = 1591
Score = 229 bits (585), Expect = 4e-61
Identities = 131/293 (44%), Positives = 177/293 (59%), Gaps = 8/293 (2%)
Frame = +2
Query: 24 ELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGF-QGEKEFLVEVLMLS 82
EL T NF + IG+G +G VY L G AVAVK+LD + + EFL +V M+S
Sbjct: 335 ELKEKTDNFGSKALIGEGSYGRVYYATLND-GNAVAVKKLDVSSEPETNNEFLTQVSMVS 511
Query: 83 LLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDL-----LPDNEPLDWNTRMRIA 137
L + N V + GYC EG+ R+L YE+ MGSL LH LDW R+RIA
Sbjct: 512 RLKNDNFVELHGYCVEGNLRVLAYEFATMGSLHDILHGRKGVQGAQPGPTLDWIQRVRIA 691
Query: 138 VGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQSYVATRVM 197
V AARGL YLH + +P++I+RD++SSN+L+ E + K++DF L+ P +TRV+
Sbjct: 692 VDAARGLEYLHEKVQPAIIHRDIRSSNVLIFEDYKAKIADFNLSNQAPDMAARLHSTRVL 871
Query: 198 GTHGYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYDETRAH-DKHLVDWARP-LFR 255
GT GY APEYA TG+LT +SD+YSFGVVLLEL+TGR+ D T + LV WA P L
Sbjct: 872 GTFGYHAPEYAMTGQLTQKSDVYSFGVVLLELLTGRKPVDHTMPRGQQSLVTWATPRLSE 1051
Query: 256 DKGNFRKLVDPHLQGHYPISGLRMALEMARMCLREDPRLRPSAGDIVLALDYL 308
DK ++ VDP L+G YP G+ +A +C++ + RP+ +V AL L
Sbjct: 1052DK--VKQCVDPKLKGEYPPKGVAKLAAVAALCVQYEAEFRPNMSIVVKALQPL 1204
>CN825263
Length = 663
Score = 228 bits (582), Expect = 1e-60
Identities = 120/217 (55%), Positives = 150/217 (68%), Gaps = 1/217 (0%)
Frame = +1
Query: 42 GFGTVYKGKLGSTGQAVAVKRLDTTGFQGEKEFLVEVLMLSLLHHPNLVSMIGYCAEGDQ 101
GFG VYKG L G+ VAVK L +G +EFL EV MLS LHH NLV +IG C E
Sbjct: 1 GFGLVYKGILND-GRDVAVKILKRDDQRGGREFLAEVEMLSRLHHRNLVKLIGICIEKQT 177
Query: 102 RLLVYEYMPMGSLESHLHDLLPDNEPLDWNTRMRIAVGAARGLNYLHHEAEPSVIYRDLK 161
R L+YE +P GS+ESHLH + PLDWN RM+IA+GAARGL YLH ++ P VI+RD K
Sbjct: 178 RCLIYELVPNGSVESHLHGADKETGPLDWNARMKIALGAARGLAYLHEDSNPCVIHRDFK 357
Query: 162 SSNILLDEGFYPKLSDFGLAKFGPTGDQSYVATRVMGTHGYCAPEYATTGKLTMRSDIYS 221
SSNILL+ F PK+SDFGLA+ +++T VMGT GY APEYA TG L ++SD+YS
Sbjct: 358 SSNILLECDFTPKVSDFGLARTALDEGNKHISTHVMGTFGYLAPEYAMTGHLLVKSDVYS 537
Query: 222 FGVVLLELITGRRAYDETR-AHDKHLVDWARPLFRDK 257
+GVVLLEL+TG + D ++ ++LV WARP+ K
Sbjct: 538 YGVVLLELLTGTKPVDLSQPPGQENLVTWARPILTSK 648
>TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, partial (64%)
Length = 969
Score = 228 bits (581), Expect = 1e-60
Identities = 116/242 (47%), Positives = 161/242 (65%), Gaps = 1/242 (0%)
Frame = +2
Query: 13 ISNKAQIFTFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGFQGEK 72
++N +IFT++EL AT F D+ +G+GGFG+VY G+ S G +AVK+L + E
Sbjct: 245 VNNSWRIFTYKELHAATGGFSDDNKLGEGGFGSVYWGRT-SDGLQIAVKKLKAMNSKAEM 421
Query: 73 EFLVEVLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLDWNT 132
EF VEV +L + H NL+ + GYC DQRL+VY+YMP SL SHLH L+W
Sbjct: 422 EFAVEVEVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQFAVEVQLNWQK 601
Query: 133 RMRIAVGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQSYV 192
RM+IA+G+A G+ YLHHE P +I+RD+K+SN+LL+ F P ++DFG AK P G S++
Sbjct: 602 RMKIAIGSAEGILYLHHEVTPHIIHRDIKASNVLLNSDFEPLVADFGFAKLIPEG-VSHM 778
Query: 193 ATRVMGTHGYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYDETRAHDKH-LVDWAR 251
TRV GT GY APEYA GK++ D+YSFG++LLEL+TGR+ ++ K + +WA
Sbjct: 779 TTRVKGTLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGVKRTITEWAE 958
Query: 252 PL 253
PL
Sbjct: 959 PL 964
>TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete
Length = 2193
Score = 223 bits (569), Expect = 3e-59
Identities = 134/305 (43%), Positives = 186/305 (60%), Gaps = 10/305 (3%)
Frame = +1
Query: 16 KAQIFTFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGFQGEKEFL 75
K+ F+++ELA AT NF + IGQGGFG VY +L G+ A+K++D Q EFL
Sbjct: 1039 KSMEFSYQELAKATNNFSLDNKIGQGGFGAVYYAEL--RGKKTAIKKMDV---QASTEFL 1203
Query: 76 VEVLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLDWNTRMR 135
E+ +L+ +HH NLV +IGYC EG LVYE++ G+L +LH EPL W++R++
Sbjct: 1204 CELKVLTHVHHLNLVRLIGYCVEGSL-FLVYEHIDNGNLGQYLHG--SGKEPLPWSSRVQ 1374
Query: 136 IAVGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQSYVATR 195
IA+ AARGL Y+H P I+RD+KS+NIL+D+ K++DFGL K G+ S + TR
Sbjct: 1375 IALDAARGLEYIHEHTVPVYIHRDVKSANILIDKNLRGKVADFGLTKLIEVGN-STLQTR 1551
Query: 196 VMGTHGYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAY---DETRAHDKHLVDWARP 252
++GT GY PEYA G ++ + D+Y+FGVVL ELI+ + A E A K LV
Sbjct: 1552 LVGTFGYMPPEYAQYGDISPKIDVYAFGVVLFELISAKNAVLKTGELVAESKGLV----A 1719
Query: 253 LFRDKGN-------FRKLVDPHLQGHYPISGLRMALEMARMCLREDPRLRPSAGDIVLAL 305
LF + N RKLVDP L +YPI + ++ R C R++P LRPS +V+AL
Sbjct: 1720 LFEEALNKSDPCDALRKLVDPRLGENYPIDSVLKIAQLGRACTRDNPLLRPSMRSLVVAL 1899
Query: 306 DYLSS 310
LSS
Sbjct: 1900 MTLSS 1914
>AV423294
Length = 459
Score = 219 bits (559), Expect = 4e-58
Identities = 100/152 (65%), Positives = 127/152 (82%)
Frame = +3
Query: 40 QGGFGTVYKGKLGSTGQAVAVKRLDTTGFQGEKEFLVEVLMLSLLHHPNLVSMIGYCAEG 99
+GGFG VY+G + Q VA+K+L+ G QG +EF+VEVL LSL HPNLV +IG+CAEG
Sbjct: 3 EGGFGKVYRGFIERINQVVAIKQLNPHGLQGIREFVVEVLTLSLADHPNLVKLIGFCAEG 182
Query: 100 DQRLLVYEYMPMGSLESHLHDLLPDNEPLDWNTRMRIAVGAARGLNYLHHEAEPSVIYRD 159
+QRLLVYEYMP+GSLESHLHD+LP +PLDWNTRM+IA GAARGL YLH++ +P VIYRD
Sbjct: 183 EQRLLVYEYMPLGSLESHLHDILPGKKPLDWNTRMKIAAGAARGLEYLHNKMKPPVIYRD 362
Query: 160 LKSSNILLDEGFYPKLSDFGLAKFGPTGDQSY 191
+K SN+L+ +G++PKLSDFGLAK GP GD+++
Sbjct: 363 IKCSNLLIGDGYHPKLSDFGLAKVGPIGDKTH 458
>TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-like
protein, partial (46%)
Length = 941
Score = 215 bits (547), Expect = 1e-56
Identities = 117/271 (43%), Positives = 172/271 (63%), Gaps = 1/271 (0%)
Frame = +3
Query: 38 IGQGGFGTVYKGKLGSTGQAVAVKRLDTTGFQGEKEFLVEVLMLSLLHHPNLVSMIGYCA 97
+G GGFG VY G++ G VA+KR + QG EF E+ MLS L H +LVS+IGYC
Sbjct: 15 LGVGGFGKVYYGEVDG-GTKVAIKRGNPLSEQGVHEFQTEIEMLSKLRHRHLVSLIGYCE 191
Query: 98 EGDQRLLVYEYMPMGSLESHLHDLLPDNEPLDWNTRMRIAVGAARGLNYLHHEAEPSVIY 157
E + +LVY++M G+L HL+ PL W R+ I +GAARGL+YLH A+ ++I+
Sbjct: 192 ENTEMILVYDHMAYGTLREHLYKT--QKPPLPWKQRLEICIGAARGLHYLHTGAKYTIIH 365
Query: 158 RDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQSYVATRVMGTHGYCAPEYATTGKLTMRS 217
RD+K++NILLDE + K+SDFGL+K GPT D ++V+T V G+ GY PEY +LT +S
Sbjct: 366 RDVKTTNILLDEKWVAKVSDFGLSKTGPTLDNTHVSTVVKGSFGYLDPEYFRRQQLTDKS 545
Query: 218 DIYSFGVVLLELITGRRAYDETRAHDK-HLVDWARPLFRDKGNFRKLVDPHLQGHYPISG 276
D+YSFGVVL E++ R A + + A ++ L +WA + +KG +++DP+L+G
Sbjct: 546 DVYSFGVVLFEILCARPALNPSLAKEQVSLAEWASHCY-NKGILDQILDPYLKGKIAPEC 722
Query: 277 LRMALEMARMCLREDPRLRPSAGDIVLALDY 307
+ E A C+ + RPS GD++ L++
Sbjct: 723 FKKFAETAMKCVSDQGIERPSMGDVLWNLEF 815
>TC19526 homologue to UP|AAQ96340 (AAQ96340) Protein kinase-like protein,
partial (37%)
Length = 432
Score = 208 bits (529), Expect = 1e-54
Identities = 100/132 (75%), Positives = 112/132 (84%)
Frame = +1
Query: 17 AQIFTFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGFQGEKEFLV 76
AQ FTFRELA ATKNFR E +G+GGFG VYKG+L STGQ VAVK+LD G QG +EFLV
Sbjct: 43 AQTFTFRELAAATKNFRPECLLGEGGFGRVYKGRLESTGQVVAVKQLDRNGLQGNREFLV 222
Query: 77 EVLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLDWNTRMRI 136
EVLMLSLLHHPNLV++IGYCA+GDQRLLVYE+MP+GSLE HLH D EPLDWNTRM+I
Sbjct: 223 EVLMLSLLHHPNLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHG---DKEPLDWNTRMKI 393
Query: 137 AVGAARGLNYLH 148
A GAA+GL YLH
Sbjct: 394 AAGAAKGLEYLH 429
>AI967315
Length = 1308
Score = 203 bits (517), Expect = 3e-53
Identities = 119/311 (38%), Positives = 179/311 (57%), Gaps = 5/311 (1%)
Frame = +1
Query: 20 FTFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTT--GFQGEKEFLVE 77
F++ EL AT F E +G+GG+ VYKG+L S G +AVKRL T + EKEFL E
Sbjct: 112 FSYEELFHATNGFSSENMVGKGGYAEVYKGRLES-GDEIAVKRLTRTCRDERKEKEFLTE 288
Query: 78 VLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLDWNTRMRIA 137
+ + + H N++ ++G C + LV+E +GS+ S +HD PLDW TR +I
Sbjct: 289 IGTIGHVCHSNVMPLLGCCIDNGL-YLVFELSTVGSVASLIHD--EKMAPLDWKTRYKIV 459
Query: 138 VGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQSYVATRVM 197
+G ARGL+YLH + +I+RD+K+SNILL E F P++SDFGLAK+ P+ + +
Sbjct: 460 LGTARGLHYLHKGCQRRIIHRDIKASNILLTEDFEPQISDFGLAKWLPSQWTHHSIAPIE 639
Query: 198 GTHGYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYDETRAHDKHLVDWARPLFRDK 257
GT G+ APEY G + ++D+++FGV LLE+I+GR+ D + + L WA+P+ K
Sbjct: 640 GTFGHLAPEYYMHGVVDEKTDVFAFGVFLLEVISGRKPVDGSH---QSLHTWAKPIL-SK 807
Query: 258 GNFRKLVDPHLQGHYPISGLRMALEMARMCLREDPRLRPSAGDIVLALDYLSSKK---YV 314
KLVDP L+G Y ++ A +C+R RP+ +++ ++ K +
Sbjct: 808 WEIEKLVDPRLEGCYDVTQFNRVAFAASLCIRASSTWRPTMSEVLEVMEEGEMDKERWKM 987
Query: 315 PKASEIVSPGE 325
PK E GE
Sbjct: 988 PKEEEEDQQGE 1020
>TC9461 similar to UP|O49840 (O49840) Protein kinase (AT2G02800/T20F6.6) ,
partial (37%)
Length = 484
Score = 193 bits (491), Expect = 3e-50
Identities = 98/163 (60%), Positives = 127/163 (77%), Gaps = 1/163 (0%)
Frame = +3
Query: 108 YMPMGSLESHLHDLLPDNEPLDWNTRMRIAVGAARGLNYLHHEAEPSVIYRDLKSSNILL 167
+MP GSLE+HL P +PL W+ RM++A+GAARGL++LH+ A+ VIYRD K+SNILL
Sbjct: 3 FMPKGSLENHLFRRGP--QPLSWSVRMKVAIGAARGLSFLHN-AKSQVIYRDFKASNILL 173
Query: 168 DEGFYPKLSDFGLAKFGPTGDQSYVATRVMGTHGYCAPEYATTGKLTMRSDIYSFGVVLL 227
D F KLSDFGLAK GPTGD+++V+T+VMGT GY APEY TG+LT +SD+YSFGVVLL
Sbjct: 174 DAEFNAKLSDFGLAKAGPTGDRTHVSTQVMGTQGYAAPEYVATGRLTAKSDVYSFGVVLL 353
Query: 228 ELITGRRAYDETRAH-DKHLVDWARPLFRDKGNFRKLVDPHLQ 269
EL++GRRA D+T A D++LVDWARP DK +++D L+
Sbjct: 354 ELLSGRRAVDKTIAGVDQNLVDWARPYLGDKRRLFRIMDSKLE 482
>TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodulation aberrant
root formation protein), complete
Length = 3308
Score = 188 bits (478), Expect = 1e-48
Identities = 113/277 (40%), Positives = 159/277 (56%), Gaps = 7/277 (2%)
Frame = +2
Query: 33 RDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGF-QGEKEFLVEVLMLSLLHHPNLVS 91
++E IG+GG G VY+G + + G VA+KRL G + + F E+ L + H N++
Sbjct: 2195 KEENIIGKGGAGIVYRGSMPN-GTDVAIKRLVGQGSGRNDYGFRAEIETLGKIRHRNIMR 2371
Query: 92 MIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLDWNTRMRIAVGAARGLNYLHHEA 151
++GY + D LL+YEYMP GSL LH + L W R +IAV AARGL Y+HH+
Sbjct: 2372 LLGYVSNKDTNLLLYEYMPNGSLGEWLHGAKGGH--LRWEMRYKIAVEAARGLCYMHHDC 2545
Query: 152 EPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQSYVATRVMGTHGYCAPEYATTG 211
P +I+RD+KS+NILLD F ++DFGLAKF S + + G++GY APEYA T
Sbjct: 2546 SPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPEYAYTL 2725
Query: 212 KLTMRSDIYSFGVVLLELITGRRAYDETRAHDKHLVDWARPLF------RDKGNFRKLVD 265
K+ +SD+YSFGVVLLELI GR+ E +V W D +VD
Sbjct: 2726 KVDEKSDVYSFGVVLLELIIGRKPVGEF-GDGVDIVGWVNKTMSELSQPSDTALVLAVVD 2902
Query: 266 PHLQGHYPISGLRMALEMARMCLREDPRLRPSAGDIV 302
P L G YP++ + +A MC++E RP+ ++V
Sbjct: 2903 PRLSG-YPLTSVIHMFNIAMMCVKEMGPARPTMREVV 3010
>TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2, complete
Length = 2946
Score = 185 bits (469), Expect = 1e-47
Identities = 122/304 (40%), Positives = 178/304 (58%), Gaps = 4/304 (1%)
Frame = +1
Query: 17 AQIFTFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGFQGEKEFLV 76
A IF F +++AT +F +G+GGFG VYKG L + GQ +AVKRL T QG +EF
Sbjct: 1627 ATIFDFSTISSATNHFSLSNKLGEGGFGPVYKGLLAN-GQEIAVKRLSNTSGQGMEEFKN 1803
Query: 77 EVLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLDWNTRMRI 136
E+ +++ L H NLV + G D+ + M + L+S L+ DWN R++I
Sbjct: 1804 EIKLIARLQHRNLVKLFGCSVHQDENSHANKKMKI-LLDSTRSKLV------DWNKRLQI 1962
Query: 137 AVGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQSYVAT-R 195
G ARGL YLH ++ +I+RDLK+SNILLD+ PK+SDFGLA+ GDQ T R
Sbjct: 1963 IDGIARGLLYLHQDSRLRIIHRDLKTSNILLDDEMNPKISDFGLARIF-IGDQVEARTKR 2139
Query: 196 VMGTHGYCAPEYATTGKLTMRSDIYSFGVVLLELITGR---RAYDETRAHDKHLVDWARP 252
VMGT+GY PEYA G +++SD++SFGV++LE+I+G+ R YD H +L+ A
Sbjct: 2140 VMGTYGYMPPEYAVHGSFSIKSDVFSFGVIVLEIISGKKIGRFYDP--HHHLNLLSHAWR 2313
Query: 253 LFRDKGNFRKLVDPHLQGHYPISGLRMALEMARMCLREDPRLRPSAGDIVLALDYLSSKK 312
L+ ++ +LVD L + + + +A +C++ P RP IVL L+ +K
Sbjct: 2314 LWIEERPL-ELVDELLDDPVIPTEILRYIHVALLCVQRRPENRPDMLSIVL---MLNGEK 2481
Query: 313 YVPK 316
+PK
Sbjct: 2482 ELPK 2493
>TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P14.15
{Arabidopsis thaliana;}, partial (37%)
Length = 1079
Score = 174 bits (440), Expect = 3e-44
Identities = 99/243 (40%), Positives = 155/243 (63%), Gaps = 7/243 (2%)
Frame = +3
Query: 77 EVLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLDWNTRMRI 136
EV +L+ + H NLV ++G+ +G++R+L+ EY+P G+L HL L + LD+N R+ I
Sbjct: 6 EVELLAKIDHRNLVKLLGFIDKGNERILITEYVPNGTLREHLDGLR--GKILDFNQRLEI 179
Query: 137 AVGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPT-GDQSYVATR 195
A+ A GL YLH AE +I+RD+KSSNILL E K++DFG A+ GP GDQ++++T+
Sbjct: 180 AIDVAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFARLGPVNGDQTHISTK 359
Query: 196 VMGTHGYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRAYDETRAHDKHL-VDWARPLF 254
V GT GY PEY T +LT +SD+YSFG++LLE++TGRR + +A D+ + + WA +
Sbjct: 360 VKGTVGYLDPEYMKTHQLTPKSDVYSFGILLLEILTGRRPVELKKAADERVTLRWAFRKY 539
Query: 255 RDKGNFRKLVDPHLQGHYPISGLRMALEMARMC---LREDPRLRPSAGDIVLAL--DYLS 309
++G+ +L+DP ++ L L+++ C +R D S G+ + A+ DYL
Sbjct: 540 -NEGSVVELMDPLMEEAVNADVLMKMLDLSFQCAAPIRTDRPNMKSVGEQLWAIRADYLK 716
Query: 310 SKK 312
S +
Sbjct: 717 SAR 725
>TC12473 similar to UP|Q9M068 (Q9M068) Protein kinase-like protein, partial
(42%)
Length = 946
Score = 170 bits (430), Expect = 4e-43
Identities = 91/182 (50%), Positives = 122/182 (67%), Gaps = 9/182 (4%)
Frame = +3
Query: 16 KAQIFTFRELATATKNFRDETFIGQGGFGTVYKG-----KLGST----GQAVAVKRLDTT 66
K ++F+F +L +ATK+F+ + IG+GGFG VYKG KL T G VAVK+L+
Sbjct: 408 KLKVFSFGDLKSATKSFKADALIGEGGFGKVYKGWLDEKKLSPTKPGSGIMVAVKKLNPE 587
Query: 67 GFQGEKEFLVEVLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNE 126
QG E+ E+ L + HPNLV ++GYC + ++ LLVYE+MP GSLE+HL + E
Sbjct: 588 SMQGFHEWQSEINFLGRISHPNLVKLLGYCRDDEEFLLVYEFMPRGSLENHL--FRRNTE 761
Query: 127 PLDWNTRMRIAVGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPT 186
L WNTR++IA+GAARGL +L H VIYRD K+SNILLD + K+S FGL KFGP+
Sbjct: 762 SLSWNTRLKIAIGAARGLAFL-HSLXKIVIYRDFKASNILLDGNYNAKISXFGLTKFGPS 938
Query: 187 GD 188
G+
Sbjct: 939 GE 944
>AV417846
Length = 429
Score = 169 bits (429), Expect = 5e-43
Identities = 85/144 (59%), Positives = 108/144 (74%), Gaps = 1/144 (0%)
Frame = +1
Query: 108 YMPMGSLESHLHDLLPDNEPLDWNTRMRIAVGAARGLNYLHHEAEPSVIYRDLKSSNILL 167
+MP GS+E+HL +P W+ R++IA+GAA+GL +LH EP VIYRD K+SNILL
Sbjct: 1 FMPKGSMENHLFRRGSYFQPFSWSLRLKIALGAAKGLAFLH-STEPKVIYRDFKTSNILL 177
Query: 168 DEGFYPKLSDFGLAKFGPTGDQSYVATRVMGTHGYCAPEYATTGKLTMRSDIYSFGVVLL 227
D + KLSDFGLA+ GP GD+S+V+TRVMGT GY APEY TG LT SD+YSFGVVLL
Sbjct: 178 DTKYSAKLSDFGLARDGPVGDKSHVSTRVMGTRGYAAPEYLATGHLTANSDVYSFGVVLL 357
Query: 228 ELITGRRAYDETRAHDKH-LVDWA 250
E+I+GRRA D+ + +H LV+WA
Sbjct: 358 EIISGRRAIDKNQLAGEHNLVEWA 429
>BP032158
Length = 555
Score = 167 bits (422), Expect = 3e-42
Identities = 87/186 (46%), Positives = 119/186 (63%)
Frame = +2
Query: 16 KAQIFTFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGFQGEKEFL 75
K F+ R++ AT NF IG+GGFG VYKG L S G +AVK+L + QG +EF+
Sbjct: 2 KTGYFSLRQIKAATNNFDPANKIGEGGFGPVYKGVL-SEGDVIAVKQLSSKSKQGNREFI 178
Query: 76 VEVLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLDWNTRMR 135
E+ M+S L HPNLV + G C EG+Q LLVYEYM SL L L+W TRM+
Sbjct: 179 NEIGMISALQHPNLVKLYGCCIEGNQLLLVYEYMENNSLARALFGNEEQKLNLNWRTRMK 358
Query: 136 IAVGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQSYVATR 195
I VG A+GL YLH E+ +++RD+K++N+LLD+ K+SDFGLAK + ++++TR
Sbjct: 359 ICVGIAKGLAYLHEESRLKIVHRDIKATNVLLDKDLKAKISDFGLAKLDEE-ENTHISTR 535
Query: 196 VMGTHG 201
+ GT G
Sbjct: 536 IAGTIG 553
>TC12704 UP|CAE45596 (CAE45596) S-receptor kinase-like protein 3, complete
Length = 2481
Score = 166 bits (419), Expect = 8e-42
Identities = 116/327 (35%), Positives = 172/327 (52%), Gaps = 25/327 (7%)
Frame = +1
Query: 17 AQIFTFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAVAVKRLDTTGFQGEKEFLV 76
A IF F +++ T +F + +G+GGFG VYKG L + GQ +AVKRL T QG +EF
Sbjct: 1408 ATIFDFSTISSTTNHFSESNKLGEGGFGPVYKGVLAN-GQEIAVKRLSNTSGQGMEEFKN 1584
Query: 77 EVLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHLHDLLPDNEPLDWNTRMRI 136
EV +++ L H NLV ++G D+ LL+YE+M SL+ + D +R+RI
Sbjct: 1585 EVKLIARLQHRNLVKLLGCSIHHDEMLLIYEFMHNRSLDYFIFD-----------SRLRI 1731
Query: 137 AVGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDFGLAKFGPTGDQSYVAT-R 195
I+RDLK+SNILLD PK+SDFGLA+ TGDQ T R
Sbjct: 1732 -------------------IHRDLKTSNILLDSEMNPKISDFGLARIF-TGDQVEAKTKR 1851
Query: 196 VMGTHGYCAPEYATTGKLTMRSDIYSFGVVLLELITGRRA--------YDETRAHDKH-- 245
VMGT+GY +PEYA G +++SD++SFGV++LE+I+G++ + +H +
Sbjct: 1852 VMGTYGYMSPEYAVHGSFSVKSDVFSFGVIVLEIISGKKIGRFCDPHHHRNLLSHSSNFA 2031
Query: 246 --LVDWARPLFRDKGNFRK------------LVDPHLQGHYPISGLRMALEMARMCLRED 291
L+ R + RK LVD L G + + + +A +C+++
Sbjct: 2032 VFLIKALRICMFENVKNRKAWRLWIEERPLELVDELLDGLAIPTEILRYIHIALLCVQQR 2211
Query: 292 PRLRPSAGDIVLALDYLSSKKYVPKAS 318
P RP +VL L+ +K +PK S
Sbjct: 2212 PEYRPDMLSVVL---MLNGEKELPKPS 2283
>TC12343 similar to UP|CDK9_SCHPO (Q96WV9) Serine/threonine protein kinase
Cdk9 (Cyclin-dependent kinase Cdk9) , partial (7%)
Length = 723
Score = 159 bits (401), Expect = 9e-40
Identities = 87/196 (44%), Positives = 121/196 (61%), Gaps = 4/196 (2%)
Frame = +3
Query: 3 SEATTAENTDISNKA----QIFTFRELATATKNFRDETFIGQGGFGTVYKGKLGSTGQAV 58
S + DI N A + F++ L ATKNF +G+GGFG V+KGKL G+ +
Sbjct: 144 SSSEAQNEDDIQNIAAKEHKTFSYETLVAATKNFHAVNKLGEGGFGPVFKGKLND-GREI 320
Query: 59 AVKRLDTTGFQGEKEFLVEVLMLSLLHHPNLVSMIGYCAEGDQRLLVYEYMPMGSLESHL 118
AVK+L QG +F+ E +L+ + H N+VS+ GYCA G ++LLVYEY+P SL+ L
Sbjct: 321 AVKKLSRRSNQGRTQFINEAKLLTRVQHRNVVSLFGYCAHGSEKLLVYEYVPRESLDKLL 500
Query: 119 HDLLPDNEPLDWNTRMRIAVGAARGLNYLHHEAEPSVIYRDLKSSNILLDEGFYPKLSDF 178
E LDW R I G ARGL YLH ++ +I+RD+K++NILLDE + PK++DF
Sbjct: 501 FRS-QKKEQLDWKRRFDIISGVARGLLYLHEDSHDCIIHRDIKAANILLDEKWVPKIADF 677
Query: 179 GLAKFGPTGDQSYVAT 194
GLA+ P DQ++V T
Sbjct: 678 GLARIFPE-DQTHVNT 722
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.136 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,740,364
Number of Sequences: 28460
Number of extensions: 74063
Number of successful extensions: 929
Number of sequences better than 10.0: 338
Number of HSP's better than 10.0 without gapping: 706
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 709
length of query: 377
length of database: 4,897,600
effective HSP length: 92
effective length of query: 285
effective length of database: 2,279,280
effective search space: 649594800
effective search space used: 649594800
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 56 (26.2 bits)
Medicago: description of AC133780.4


