

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133779.6 - phase: 0
(1061 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
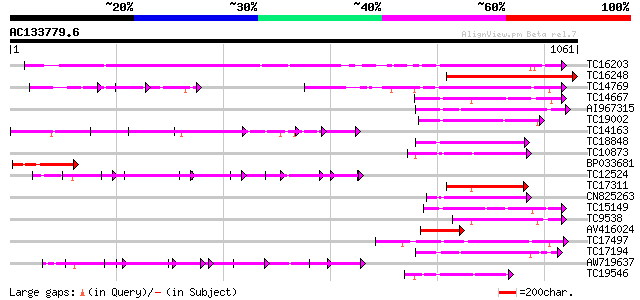
Score E
Sequences producing significant alignments: (bits) Value
TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodula... 442 e-124
TC16248 similar to UP|BAC87845 (BAC87845) Leucine-rich repeat re... 385 e-107
TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete 181 7e-46
TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, com... 162 2e-40
AI967315 160 1e-39
TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866)... 157 8e-39
TC14163 157 1e-38
TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, pa... 156 2e-38
TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partia... 152 2e-37
BP033681 151 5e-37
TC12524 similar to UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kina... 148 4e-36
TC17311 similar to UP|Q8K2Y2 (Q8K2Y2) Receptor-interacting serin... 143 2e-34
CN825263 142 4e-34
TC15149 similar to UP|Q9LVI6 (Q9LVI6) Probable receptor-like pro... 141 6e-34
TC9538 similar to UP|RLK5_ARATH (P47735) Receptor-like protein k... 141 6e-34
AV416024 139 2e-33
TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2,... 137 1e-32
TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete 135 4e-32
AW719637 134 6e-32
TC19546 similar to UP|Q40543 (Q40543) Protein-serine/threonine k... 129 2e-30
>TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodulation aberrant
root formation protein), complete
Length = 3308
Score = 442 bits (1137), Expect = e-124
Identities = 320/1034 (30%), Positives = 506/1034 (47%), Gaps = 20/1034 (1%)
Frame = +2
Query: 28 ALLKWKASFDNQSQS--ILSTWKNTTN---PCSKWRGIECDKSNLISTIDLANLGLKGTL 82
ALLK K S L WK +T+ CS + G+ CD+ NL
Sbjct: 203 ALLKLKESMKGAKAKHHALEDWKFSTSLSAHCS-FSGVTCDQ----------NL------ 331
Query: 83 HSLTFSSFPNLITLNIYNNHFYGTIPPQIGNLSRINTLNFSKNPIIGSIPQEMYTLRSLK 142
++ LN+ +G +PP+IG L ++ L S N + +P ++ +L SLK
Sbjct: 332 ---------RVVALNVTLVPLFGHLPPEIGLLEKLENLTISMNNLTDQLPSDLASLTSLK 484
Query: 143 GLDFFFCTLSGEIDKSIG-NLTNLSYLDLGGNNFSGGPIPPEIGKLKKLRYLAITQGSLV 201
L+ SG+ +I +T L LD N+FSG P+P EI KL+KL+YL +
Sbjct: 485 VLNISHNLFSGQFPGNITVGMTELEALDAYDNSFSG-PLPEEIVKLEKLKYLHLAGNYFS 661
Query: 202 GSIPQEIGLLTNLTYIDLSNNFLSGVIPETIGNMSKLNQLMFANNTKLYGPIPHSLWNMS 261
G+IP+ +L ++ L+ N L+G +PE++ + L +L + G IP + +M
Sbjct: 662 GTIPESYSEFQSLEFLGLNANSLTGRVPESLAKLKTLKELHLGYSNAYEGGIPPAFGSME 841
Query: 262 SLTLIYLYNMSLSGSIPDSVQNLINLDVLALYMNNLSGFIPSTIGNLKNLTLLLLRNNRL 321
+L L+ + N +L+G IP S+ NL L L + MNNL+G IP + ++ +L L L N L
Sbjct: 842 NLRLLEMANCNLTGEIPPSLGNLTKLHSLFVQMNNLTGTIPPELSSMMSLMSLDLSINDL 1021
Query: 322 SGSIPASIGNLINLKYFSVQVNNLTGTIPATIGNLKQLIVFEVASNKLYGRIPNGLYNIT 381
+G IP S L NL + N G++P+ IG+L L +V N +P+ L
Sbjct: 1022 TGEIPESFSKLKNLTLMNFFQNKFRGSLPSFIGDLPNLETLQVWENNFSFVLPHNLGGNG 1201
Query: 382 NWYSFVVSENDFVGHLPSQMCTGGSLKYLSAFHNRFTGPVPTSLKSCSSIERIRIEGNQI 441
+ F V++N G +P +C G LK N F GP+P + C S+ +IR+ N +
Sbjct: 1202 RFLYFDVTKNHLTGLIPPDLCKSGRLKTFIITDNFFRGPIPKGIGECRSLTKIRVANNFL 1381
Query: 442 EGDIAEDFGVYPNLRYVDLSDNKFHGHISPNWGKSLDLETFMISNTNISGGIPLDFIGLT 501
+G + P++ +LS+N+ +G + P+ L T +SN +G IP L
Sbjct: 1382 DGPVPPGVFQLPSVTITELSNNRLNGEL-PSVISGESLGTLTLSNNLFTGKIPAAMKNLR 1558
Query: 502 KLGRLHLSSNQLTGKLPKEILGGMKSLLYLKISNNHFTDSIPTEIGLLQRLEELDLGGNE 561
L L L +N+ G++P + + L + IS N+ T IPT I L +DL N
Sbjct: 1559 ALQSLSLDANEFIGEIPGGVF-EIPMLTKVNISGNNLTGPIPTTITHRASLTAVDLSRNN 1735
Query: 562 LSGTIPNEVAELPKLRMLNLSRNRIEGRIPS--TFDSALASIDLSGNRLNGNIPTSLGFL 619
L+G +P + L L +LNLSRN I G +P F ++L ++DLS N G +PT FL
Sbjct: 1736 LAGEVPKGMKNLMDLSILNLSRNEISGPVPDEIRFMTSLTTLDLSSNNFTGTVPTGGQFL 1915
Query: 620 VQLSMLNLSHNMLSGTIPSTFSMSLDFVNISDNQLDGPLPENPAFLRAPFESFKNNKGLC 679
V TF+ + + P + ++S + +
Sbjct: 1916 V-------------FNYDKTFAGNPNLC----------FPHRASCPSVLYDSLRKTRAKT 2026
Query: 680 GNITGLVPCATSQIHSRKSKNILQSVFIALGALILVLSGVGISMYVFFRRKKPNEEIQTE 739
+ +V + IAL +L+ V ++++V +R+ +
Sbjct: 2027 ARVRAIV------------------IGIALATAVLL---VAVTVHVVRKRRLHRAQAWKL 2143
Query: 740 EEVQKGVLFSIWSHDGKMMFENIIEATENFDDKYLIGVGSQGNVYKAELPTGLVVAVKKL 799
Q+ ++ E+++E + ++ +IG G G VY+ +P G VA+K+L
Sbjct: 2144 TAFQR----------LEIKAEDVVECLK---EENIIGKGGAGIVYRGSMPNGTDVAIKRL 2284
Query: 800 HLVRDEEMSFFSSKSFTSEIETLTGIKHRNIIKLHGFCSHSKFSFLVYKFMEGGSLDQIL 859
+ S + F +EIETL I+HRNI++L G+ S+ + L+Y++M GSL + L
Sbjct: 2285 ----VGQGSGRNDYGFRAEIETLGKIRHRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWL 2452
Query: 860 NNEKQAIAFDWEKRVNVVKGVANALSYLHHDCSPPIIHRDISSKNILLNLDYEAHVSDFG 919
+ K WE R + A L Y+HHDCSP IIHRD+ S NILL+ D+EAHV+DFG
Sbjct: 2453 HGAKGG-HLRWEMRYKIAVEAARGLCYMHHDCSPLIIHRDVKSNNILLDADFEAHVADFG 2629
Query: 920 TAKFLKPD--LHSWTQFAGTFGYAAPELSQTMEVNEKCDVYSFGVLALEIIIGKHP---- 973
AKFL S + AG++GY APE + T++V+EK DVYSFGV+ LE+IIG+ P
Sbjct: 2630 LAKFLYDPGASQSMSSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELIIGRKPVGEF 2809
Query: 974 GDLISLF------LSPSTRPTANDMLLTEVLDQRPQKVIKPIDEEVILIAKLAFSCLNQV 1027
GD + + +S ++P+ ++L V P+ P+ VI + +A C+ ++
Sbjct: 2810 GDGVDIVGWVNKTMSELSQPSDTALVLAVV---DPRLSGYPL-TSVIHMFNIAMMCVKEM 2977
Query: 1028 PRSRPTMDQVCKML 1041
+RPTM +V ML
Sbjct: 2978 GPARPTMREVVHML 3019
>TC16248 similar to UP|BAC87845 (BAC87845) Leucine-rich repeat receptor-like
protein kinase 1, partial (17%)
Length = 866
Score = 385 bits (989), Expect = e-107
Identities = 187/247 (75%), Positives = 212/247 (85%), Gaps = 2/247 (0%)
Frame = +2
Query: 817 SEIETLTGIKHRNIIKLHGFCSHSKFSFLVYKFMEGGSLDQILNNEKQAIAFDWEKRVNV 876
SEI+TLT I+HRN+IKLHGFC HSKFSFLVY+F+EGGSLDQ+LN++ QA AF WEKRVNV
Sbjct: 2 SEIQTLTEIRHRNVIKLHGFCLHSKFSFLVYEFLEGGSLDQVLNSDTQAAAFHWEKRVNV 181
Query: 877 VKGVANALSYLHHDCSPPIIHRDISSKNILLNLDYEAHVSDFGTAKFLKPDLHSWTQFAG 936
VKGVANALSY+HHDCSPPIIHRDISSKN+LL+L YEAHVSDFGTAKFLKP H+WT FAG
Sbjct: 182 VKGVANALSYMHHDCSPPIIHRDISSKNVLLDLQYEAHVSDFGTAKFLKPGSHTWTAFAG 361
Query: 937 TFGYAAPELSQTMEVNEKCDVYSFGVLALEIIIGKHPGDLISLFLSPSTRPTANDMLLTE 996
TFGYAAPEL+QTM+VNEKCDVYSFGV ALEII+G HPGDLIS +SPST P ND+LL +
Sbjct: 362 TFGYAAPELAQTMQVNEKCDVYSFGVFALEIIMGMHPGDLISSLMSPSTVPMVNDLLLID 541
Query: 997 VLDQRPQKVIKPIDEEVILIAKLAFSCLNQVPRSRPTMDQVCKMLGAGKSPLE--NQFHT 1054
+LDQRP +V KPIDEEVILIA LA +CL + P SRPTMDQV K L GK PL QF
Sbjct: 542 ILDQRPHQVEKPIDEEVILIASLALACLRKNPHSRPTMDQVSKALVLGKPPLALGYQFPM 721
Query: 1055 IKLGQLH 1061
+++GQLH
Sbjct: 722 VRVGQLH 742
>TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete
Length = 3495
Score = 181 bits (458), Expect = 7e-46
Identities = 151/515 (29%), Positives = 234/515 (45%), Gaps = 25/515 (4%)
Frame = +3
Query: 552 LEELDLGGNELSGTIPNEVAELPKLRMLNLSRNRIEGRIPS-TFDSALASIDLSGNRLNG 610
+ +LDL + L G IP+ +AE+ L LN+S N +G +PS S L S+DLS N L G
Sbjct: 1599 ITKLDLSSSNLKGLIPSSIAEMTNLETLNISHNSFDGSVPSFPLSSLLISVDLSYNDLMG 1778
Query: 611 NIPTSLGFLVQLSMLNLSHNMLSGTIPSTFSMSLDFVNISDNQLDGPLPENPAFLRAPFE 670
+P S+ L L L N + PE+PA + +
Sbjct: 1779 KLPESIVKLPHLKSLYFGCN------------------------EHMSPEDPANMNSSL- 1883
Query: 671 SFKNNKGLCGNITGLVPCATSQIHSRKSKNILQSVFIALGAL----ILVLSGVGISMYVF 726
+ G C K K I +GA+ +L+ G+
Sbjct: 1884 -INTDYGRC-----------------KGKESRFGQVIVIGAITCGSLLITLAFGVLFVCR 2009
Query: 727 FRRKK-PNEEIQTEE-EVQKGVLFSIWSHDG---------KMMFENIIEATENFDDKYLI 775
+R+K P E ++ ++ ++FS+ S D E I ATE + K LI
Sbjct: 2010 YRQKLIPWEGFAGKKYPMETNIIFSLPSKDDFFIKSVSIQAFTLEYIEVATERY--KTLI 2183
Query: 776 GVGSQGNVYKAELPTGLVVAVKKLHLVRDEEMSFFSSKSFTSEIETLTGIKHRNIIKLHG 835
G G G+VY+ L G VAVK VR S ++ F +E+ L+ I+H N++ L G
Sbjct: 2184 GEGGFGSVYRGTLNDGQEVAVK----VR-SSTSTQGTREFDNELNLLSAIQHENLVPLLG 2348
Query: 836 FCSHSKFSFLVYKFMEGGSL-DQILNNEKQAIAFDWEKRVNVVKGVANALSYLHHDCSPP 894
+C+ S LVY FM GSL D++ + DW R+++ G A L+YLH
Sbjct: 2349 YCNESDQQILVYPFMSNGSLQDRLYGEPAKRKILDWPTRLSIALGAARGLAYLHTFPGRS 2528
Query: 895 IIHRDISSKNILLNLDYEAHVSDFGTAKFLKPDLHSWT--QFAGTFGYAAPELSQTMEVN 952
+IHRDI S NILL+ A V+DFG +K+ + S+ + GT GY PE +T +++
Sbjct: 2529 VIHRDIKSSNILLDHSMCAKVADFGFSKYAPQEGDSYVSLEVRGTAGYLDPEYYKTQQLS 2708
Query: 953 EKCDVYSFGVLALEIIIGKHPGDLISLFLSPSTRPTANDMLLTEVLDQRPQKVIKP---- 1008
EK DV+SFGV+ LEI+ G+ P ++ P T + + + + +++ P
Sbjct: 2709 EKSDVFSFGVVLLEIVSGREPLNI----KRPRTEWSLVEWATPYIRGSKVDEIVDPGIKG 2876
Query: 1009 --IDEEVILIAKLAFSCLNQVPRSRPTMDQVCKML 1041
E + + ++A CL RP+M + + L
Sbjct: 2877 GYHAEAMWRVVEVALQCLEPFSTYRPSMVAIVREL 2981
Score = 49.3 bits (116), Expect = 3e-06
Identities = 40/140 (28%), Positives = 58/140 (40%), Gaps = 4/140 (2%)
Frame = +3
Query: 38 NQSQSILSTWKNTTNPCSKWRGIECDKSN---LISTIDLANLGLKGTLHSLTFSSFPNLI 94
N L +W W+GI CD SN +I+ +DL++ LKG
Sbjct: 1500 NSGNRALESWSGDPCILLPWKGIACDGSNGSSVITKLDLSSSNLKGL------------- 1640
Query: 95 TLNIYNNHFYGTIPPQIGNLSRINTLNFSKNPIIGSIPQEMYTLRSLK-GLDFFFCTLSG 153
IP I ++ + TLN S N GS+P + L SL +D + L G
Sbjct: 1641 ------------IPSSIAEMTNLETLNISHNSFDGSVPS--FPLSSLLISVDLSYNDLMG 1778
Query: 154 EIDKSIGNLTNLSYLDLGGN 173
++ +SI L +L L G N
Sbjct: 1779 KLPESIVKLPHLKSLYFGCN 1838
Score = 44.7 bits (104), Expect = 8e-05
Identities = 34/98 (34%), Positives = 48/98 (48%)
Frame = +3
Query: 165 LSYLDLGGNNFSGGPIPPEIGKLKKLRYLAITQGSLVGSIPQEIGLLTNLTYIDLSNNFL 224
++ LDL +N G IP I ++ L L I+ S GS+P L + L +DLS N L
Sbjct: 1599 ITKLDLSSSNLKG-LIPSSIAEMTNLETLNISHNSFDGSVPS-FPLSSLLISVDLSYNDL 1772
Query: 225 SGVIPETIGNMSKLNQLMFANNTKLYGPIPHSLWNMSS 262
G +PE+I + L L F N + P NM+S
Sbjct: 1773 MGKLPESIVKLPHLKSLYFGCNEHM---SPEDPANMNS 1877
Score = 43.5 bits (101), Expect = 2e-04
Identities = 43/165 (26%), Positives = 71/165 (42%), Gaps = 4/165 (2%)
Frame = +3
Query: 199 SLVGSIPQEIGLLTNLTYIDLSNNFLSGVIPETIGNMSKLNQLMFANNTKLYGPIPHSLW 258
+LV + E G L N I ++ +G + K+ + + N+ G W
Sbjct: 1359 TLVKASKSEFGPLLNAYEILQVRPWIEETNQTDVGVIQKMREELLLQNS---GNRALESW 1529
Query: 259 NMSSLTLIYLYNMSLSGSIPDSVQNLINLDVLALYMNNLSGFIPSTIGNLKNLTLLLLRN 318
+ L+ ++ GS SV + L L +NL G IPS+I + NL L + +
Sbjct: 1530 SGDPCILLPWKGIACDGSNGSSV-----ITKLDLSSSNLKGLIPSSIAEMTNLETLNISH 1694
Query: 319 NRLSGSIP----ASIGNLINLKYFSVQVNNLTGTIPATIGNLKQL 359
N GS+P +S+ ++L Y N+L G +P +I L L
Sbjct: 1695 NSFDGSVPSFPLSSLLISVDLSY-----NDLMGKLPESIVKLPHL 1814
Score = 37.7 bits (86), Expect = 0.010
Identities = 30/91 (32%), Positives = 45/91 (48%), Gaps = 2/91 (2%)
Frame = +3
Query: 458 VDLSDNKFHGHISPNWGKSLDLETFMISNTNISGGIPLDFIGLTKLGRLHLSSNQLTGKL 517
+DLS + G I + + +LET IS+ + G +P F + L + LS N L GKL
Sbjct: 1608 LDLSSSNLKGLIPSSIAEMTNLETLNISHNSFDGSVP-SFPLSSLLISVDLSYNDLMGKL 1784
Query: 518 PKEI--LGGMKSLLYLKISNNHFTDSIPTEI 546
P+ I L +KSL + N H + P +
Sbjct: 1785 PESIVKLPHLKSLYF--GCNEHMSPEDPANM 1871
Score = 33.5 bits (75), Expect = 0.18
Identities = 23/84 (27%), Positives = 41/84 (48%)
Frame = +3
Query: 388 VSENDFVGHLPSQMCTGGSLKYLSAFHNRFTGPVPTSLKSCSSIERIRIEGNQIEGDIAE 447
+S ++ G +PS + +L+ L+ HN F G VP S S + + + N + G + E
Sbjct: 1614 LSSSNLKGLIPSSIAEMTNLETLNISHNSFDGSVP-SFPLSSLLISVDLSYNDLMGKLPE 1790
Query: 448 DFGVYPNLRYVDLSDNKFHGHISP 471
P+L+ + N+ H+SP
Sbjct: 1791 SIVKLPHLKSLYFGCNE---HMSP 1853
>TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, complete
Length = 1591
Score = 162 bits (411), Expect = 2e-40
Identities = 107/304 (35%), Positives = 153/304 (50%), Gaps = 19/304 (6%)
Frame = +2
Query: 757 MMFENIIEATENFDDKYLIGVGSQGNVYKAELPTGLVVAVKKLHLVRDEEMSFFSSKSFT 816
+ + + E T+NF K LIG GS G VY A L G VAVKKL + + E ++ F
Sbjct: 323 LSLDELKEKTDNFGSKALIGEGSYGRVYYATLNDGNAVAVKKLDVSSEPE----TNNEFL 490
Query: 817 SEIETLTGIKHRNIIKLHGFCSHSKFSFLVYKFMEGGSLDQILNNEK------QAIAFDW 870
+++ ++ +K+ N ++LHG+C L Y+F GSL IL+ K DW
Sbjct: 491 TQVSMVSRLKNDNFVELHGYCVEGNLRVLAYEFATMGSLHDILHGRKGVQGAQPGPTLDW 670
Query: 871 EKRVNVVKGVANALSYLHHDCSPPIIHRDISSKNILLNLDYEAHVSDF---GTAKFLKPD 927
+RV + A L YLH P IIHRDI S N+L+ DY+A ++DF A +
Sbjct: 671 IQRVRIAVDAARGLEYLHEKVQPAIIHRDIRSSNVLIFEDYKAKIADFNLSNQAPDMAAR 850
Query: 928 LHSWTQFAGTFGYAAPELSQTMEVNEKCDVYSFGVLALEIIIGKHPGDLISLFLSPSTRP 987
LHS T+ GTFGY APE + T ++ +K DVYSFGV+ LE++ G+ P D T P
Sbjct: 851 LHS-TRVLGTFGYHAPEYAMTGQLTQKSDVYSFGVVLLELLTGRKPVD--------HTMP 1003
Query: 988 TANDMLLTEVLDQRPQ-KVIKPIDEE---------VILIAKLAFSCLNQVPRSRPTMDQV 1037
L+T + + KV + +D + V +A +A C+ RP M V
Sbjct: 1004 RGQQSLVTWATPRLSEDKVKQCVDPKLKGEYPPKGVAKLAAVAALCVQYEAEFRPNMSIV 1183
Query: 1038 CKML 1041
K L
Sbjct: 1184 VKAL 1195
>AI967315
Length = 1308
Score = 160 bits (405), Expect = 1e-39
Identities = 103/297 (34%), Positives = 154/297 (51%), Gaps = 6/297 (2%)
Frame = +1
Query: 759 FENIIEATENFDDKYLIGVGSQGNVYKAELPTGLVVAVKKL-HLVRDEEMSFFSSKSFTS 817
+E + AT F + ++G G VYK L +G +AVK+L RDE K F +
Sbjct: 118 YEELFHATNGFSSENMVGKGGYAEVYKGRLESGDEIAVKRLTRTCRDER----KEKEFLT 285
Query: 818 EIETLTGIKHRNIIKLHGFCSHSKFSFLVYKFMEGGSLDQILNNEKQAIAFDWEKRVNVV 877
EI T+ + H N++ L G C + +LV++ GS+ ++++EK A DW+ R +V
Sbjct: 286 EIGTIGHVCHSNVMPLLGCCIDNGL-YLVFELSTVGSVASLIHDEKMA-PLDWKTRYKIV 459
Query: 878 KGVANALSYLHHDCSPPIIHRDISSKNILLNLDYEAHVSDFGTAKFLKPDL--HSWTQFA 935
G A L YLH C IIHRDI + NILL D+E +SDFG AK+L HS
Sbjct: 460 LGTARGLHYLHKGCQRRIIHRDIKASNILLTEDFEPQISDFGLAKWLPSQWTHHSIAPIE 639
Query: 936 GTFGYAAPELSQTMEVNEKCDVYSFGVLALEIIIGKHPGDLISLFLSPSTRPTANDMLLT 995
GTFG+ APE V+EK DV++FGV LE+I G+ P D L +P + +
Sbjct: 640 GTFGHLAPEYYMHGVVDEKTDVFAFGVFLLEVISGRKPVDGSHQSLHTWAKPILSKWEIE 819
Query: 996 EVLDQRPQKVIKPIDEEVILIAKLAFS---CLNQVPRSRPTMDQVCKMLGAGKSPLE 1049
+++D R + +V ++AF+ C+ RPTM +V +++ G+ E
Sbjct: 820 KLVDPRLEGCY-----DVTQFNRVAFAASLCIRASSTWRPTMSEVLEVMEEGEMDKE 975
>TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866), partial
(48%)
Length = 780
Score = 157 bits (397), Expect = 8e-39
Identities = 91/244 (37%), Positives = 136/244 (55%), Gaps = 7/244 (2%)
Frame = +1
Query: 765 ATENFDDKYLIGVGSQGNVYKAELPTGLVVAVKKLHLVRDEEMSFFSSKSFTSEIETLTG 824
AT NF+ +G G G+VY +L G +AVK+L + ++ + F E+E L
Sbjct: 52 ATNNFNYDNKLGEGGFGSVYWGQLWDGSQIAVKRLKVWSNK-----ADMEFAVEVEILAR 216
Query: 825 IKHRNIIKLHGFCSHSKFSFLVYKFMEGGSLDQILNNEKQA-IAFDWEKRVNVVKGVANA 883
++H+N++ L G+C+ + +VY +M SL L+ + + DW +R+N+ G A
Sbjct: 217 VRHKNLLSLRGYCAEGQERLIVYDYMPNLSLLSHLHGQHSSECLLDWNRRMNIAIGSAEG 396
Query: 884 LSYLHHDCSPPIIHRDISSKNILLNLDYEAHVSDFGTAKFLKPD--LHSWTQFAGTFGYA 941
+ YLHH +P IIHRDI + N+LL+ D++A V+DFG AK + PD H T+ GT GY
Sbjct: 397 IVYLHHQATPHIIHRDIKASNVLLDSDFQARVADFGFAKLI-PDGATHVTTRVKGTLGYL 573
Query: 942 APELSQTMEVNEKCDVYSFGVLALEIIIGKHPGDLISLFLSPSTR----PTANDMLLTEV 997
APE + + NE CDV+SFG+L LE+ GK P + +S + S P A TE
Sbjct: 574 APEYAMLGKANECCDVFSFGILLLELASGKKPLEKLSSTVKRSINDWALPLACAKKFTEF 753
Query: 998 LDQR 1001
D R
Sbjct: 754 ADPR 765
>TC14163
Length = 1712
Score = 157 bits (396), Expect = 1e-38
Identities = 118/380 (31%), Positives = 181/380 (47%), Gaps = 8/380 (2%)
Frame = +1
Query: 222 NFLSGVIPETIGNMSKLNQLMFANNTKLYGPIPHSLWNMSSLTLIYLYNMSLSGSIPDSV 281
+F SG I ++ + L+ N + GP P L+ + L IY+ N LSG IP+++
Sbjct: 370 SFFSGTISPSLSKIKNLDGFYLLNLKNISGPFPGFLFKLPKLQFIYIENNQLSGRIPENI 549
Query: 282 QNLINLDVLALYMNNLSGFIPSTIGNLKNLTLLLLRNNRLSGSIPASIGNLINLKYFSVQ 341
NL LDVL+L N +G IPS++G L +LT L L NN L+G+IPA+I L NL Y S++
Sbjct: 550 GNLTRLDVLSLTGNRFTGTIPSSVGGLTHLTQLQLGNNSLTGTIPATIARLKNLTYLSLE 729
Query: 342 VNNLTGTIPATIGNLKQLIVFEVASNKLYGRIPNGLYNITNWYSFV-VSENDFVGHLPSQ 400
N +G IP + L + ++ NK G+IP + + ++ + N G +P
Sbjct: 730 GNQFSGAIPDFFSSFTDLGILRLSRNKFSGKIPASISTLAPKLRYLELGHNQLSGKIPDF 909
Query: 401 MCTGGSLKYLSAFHNRFTGPVPTSLKSCSSIERIRIEGNQIEGDIAEDFGVYPNLRYVDL 460
+ +L L NRF+G VP S K+ + I + + N + E + +DL
Sbjct: 910 LGKFRALDTLDLSSNRFSGTVPASFKNLTKIFNLNLANNLLVDPFPE--MNVKGIESLDL 1083
Query: 461 SDNKFHGHISPNWGKSLDLETFMISNTNISGGIPLDFIGLTK---LGRLHLSSNQLTGKL 517
S+N FH + P W S + F + + LD + + LS N+++G
Sbjct: 1084SNNMFHLNQIPKWVTSSPI-IFSLKLARCGIKMKLDDWKPAETYFYDFIDLSGNEISG-- 1254
Query: 518 PKEILGGMKSLLYL----KISNNHFTDSIPTEIGLLQRLEELDLGGNELSGTIPNEVAEL 573
+G + S YL N D + G +R + LDL N + G +P VA L
Sbjct: 1255--SAVGLVNSTEYLVGFWGSGNKLKFDFERLKFG--ERFKYLDLSHNLVFGKVPKSVAGL 1422
Query: 574 PKLRMLNLSRNRIEGRIPST 593
K LN+S N + G IP T
Sbjct: 1423EK---LNVSYNHLCGEIPKT 1473
Score = 140 bits (352), Expect = 1e-33
Identities = 117/400 (29%), Positives = 181/400 (45%), Gaps = 8/400 (2%)
Frame = +1
Query: 152 SGEIDKSIGNLTNLSYLDLGGNNFSGGPIPPEIGKLKKLRYLAITQGSLVGSIPQEIGLL 211
SG I S+ + NL L GP P + KL KL+++ I L G IP+ IG L
Sbjct: 379 SGTISPSLSKIKNLDGFYLLNLKNISGPFPGFLFKLPKLQFIYIENNQLSGRIPENIGNL 558
Query: 212 TNLTYIDLSNNFLSGVIPETIGNMSKLNQLMFANNTKLYGPIPHSLWNMSSLTLIYLYNM 271
T L + L+ N +G IP ++G ++ L QL NN
Sbjct: 559 TRLDVLSLTGNRFTGTIPSSVGGLTHLTQLQLGNN------------------------- 663
Query: 272 SLSGSIPDSVQNLINLDVLALYMNNLSGFIPSTIGNLKNLTLLLLRNNRLSGSIPASIGN 331
SL+G+IP ++ L NL L+L N SG IP + +L +L L N+ SG IPASI
Sbjct: 664 SLTGTIPATIARLKNLTYLSLEGNQFSGAIPDFFSSFTDLGILRLSRNKFSGKIPASIST 843
Query: 332 LI-NLKYFSVQVNNLTGTIPATIGNLKQLIVFEVASNKLYGRIPNGLYNITNWYSFVVSE 390
L L+Y + N L+G IP +G + L +++SN+ G +P N+T ++ ++
Sbjct: 844 LAPKLRYLELGHNQLSGKIPDFLGKFRALDTLDLSSNRFSGTVPASFKNLTKIFNLNLAN 1023
Query: 391 NDFVGHLPSQMCTGGSLKYLSAFHNRF-TGPVPTSLKSCSSIERIRIE--GNQIEGDIAE 447
N V P G ++ L +N F +P + S I +++ G +++ D +
Sbjct: 1024NLLVDPFPEMNVKG--IESLDLSNNMFHLNQIPKWVTSSPIIFSLKLARCGIKMKLDDWK 1197
Query: 448 DFGVYPNLRYVDLSDNKFHGHISPNWGKSLDLETFMISNTNISGGIPLDFIGLTKLGR-- 505
Y ++DLS N+ IS + ++ +++ + DF L K G
Sbjct: 1198PAETY-FYDFIDLSGNE----ISGSAVGLVNSTEYLVGFWGSGNKLKFDFERL-KFGERF 1359
Query: 506 --LHLSSNQLTGKLPKEILGGMKSLLYLKISNNHFTDSIP 543
L LS N + GK+PK + G L L +S NH IP
Sbjct: 1360KYLDLSHNLVFGKVPKSVAG----LEKLNVSYNHLCGEIP 1467
Score = 127 bits (320), Expect = 7e-30
Identities = 99/359 (27%), Positives = 153/359 (42%), Gaps = 11/359 (3%)
Frame = +1
Query: 309 KNLTLLLLRNNR------LSGSIPASIGNLINLK-YFSVQVNNLTGTIPATIGNLKQLIV 361
K +T L L N SG+I S+ + NL ++ + + N++G P + L +L
Sbjct: 322 KRVTSLYLSGNPENPKSFFSGTISPSLSKIKNLDGFYLLNLKNISGPFPGFLFKLPKLQF 501
Query: 362 FEVASNKLYGRIPNGLYNITNWYSFVVSENDFVGHLPSQMCTGGSLKYLSAFHNRFTGPV 421
+ +N+L GRIP + N+T L LS NRFTG +
Sbjct: 502 IYIENNQLSGRIPENIGNLTR------------------------LDVLSLTGNRFTGTI 609
Query: 422 PTSLKSCSSIERIRIEGNQIEGDIAEDFGVYPNLRYVDLSDNKFHGHISPNWGKSLDLET 481
P+S+ + + ++++ N + G I NL Y+ L N+F
Sbjct: 610 PSSVGGLTHLTQLQLGNNSLTGTIPATIARLKNLTYLSLEGNQF---------------- 741
Query: 482 FMISNTNISGGIPLDFIGLTKLGRLHLSSNQLTGKLPKEILGGMKSLLYLKISNNHFTDS 541
SG IP F T LG L LS N+ +GK+P I L YL++ +N +
Sbjct: 742 --------SGAIPDFFSSFTDLGILRLSRNKFSGKIPASISTLAPKLRYLELGHNQLSGK 897
Query: 542 IPTEIGLLQRLEELDLGGNELSGTIPNEVAELPKLRMLNLSRNRIEGRIPSTFDSALASI 601
IP +G + L+ LDL N SGT+P L K+ LNL+ N + P + S+
Sbjct: 898 IPDFLGKFRALDTLDLSSNRFSGTVPASFKNLTKIFNLNLANNLLVDPFPEMNVKGIESL 1077
Query: 602 DLSGNRLNGN-IP---TSLGFLVQLSMLNLSHNMLSGTIPSTFSMSLDFVNISDNQLDG 656
DLS N + N IP TS + L + M + DF+++S N++ G
Sbjct: 1078DLSNNMFHLNQIPKWVTSSPIIFSLKLARCGIKMKLDDWKPAETYFYDFIDLSGNEISG 1254
Score = 114 bits (286), Expect = 6e-26
Identities = 120/457 (26%), Positives = 195/457 (42%), Gaps = 13/457 (2%)
Frame = +1
Query: 2 VLPTFIIMILCVLPTLSVAEDSEAKLALLKWKASFDNQSQSILSTWKNTTNPCSKWRGIE 61
+L F I L L + + + L+ +K+ + IL +W T+ C+ W+G+
Sbjct: 130 LLILFAIFTLLHLKANGAKCNVDDEAGLMGFKSGIKSDPSGILKSWIPGTDCCT-WQGVT 306
Query: 62 C--DKSNLISTIDLANLG-----LKGTLHSLTFSSFPNLITLNIYN-NHFYGTIPPQIGN 113
C D + S N GT+ S + S NL + N + G P +
Sbjct: 307 CLFDDKRVTSLYLSGNPENPKSFFSGTI-SPSLSKIKNLDGFYLLNLKNISGPFPGFLFK 483
Query: 114 LSRINTLNFSKNPIIGSIPQEMYTLRSLKGLDFFFCTLSGEIDKSIGNLTNLSYLDLGGN 173
L ++ + N + G IP+ + L L L +G I S+G LT+L+ L LG N
Sbjct: 484 LPKLQFIYIENNQLSGRIPENIGNLTRLDVLSLTGNRFTGTIPSSVGGLTHLTQLQLGNN 663
Query: 174 NFSGGPIPPEIGKLKKLRYLAITQGSLVGSIPQEIGLLTNLTYIDLSNNFLSGVIPETIG 233
+ + G IP I +LK L YL++ G+IP T+L + LS N SG IP +I
Sbjct: 664 SLT-GTIPATIARLKNLTYLSLEGNQFSGAIPDFFSSFTDLGILRLSRNKFSGKIPASIS 840
Query: 234 NMSKLNQLMFANNTKLYGPIPHSLWNMSSLTLIYLYNMSLSGSIPDSVQNLINLDVLALY 293
++ + + + +L G IP L +L + L + SG++P S +NL + L L
Sbjct: 841 TLAPKLRYLELGHNQLSGKIPDFLGKFRALDTLDLSSNRFSGTVPASFKNLTKIFNLNLA 1020
Query: 294 MNNLSGFIPSTIGNLKNLTLLLLRNNRLS-GSIPASIGN---LINLKYFSVQVN-NLTGT 348
N L P N+K + L L NN IP + + + +LK + L
Sbjct: 1021NNLLVDPFPEM--NVKGIESLDLSNNMFHLNQIPKWVTSSPIIFSLKLARCGIKMKLDDW 1194
Query: 349 IPATIGNLKQLIVFEVASNKLYGRIPNGLYNITNWYSFVVSENDFVGHLPSQMCTGGSLK 408
PA +++ N++ G GL N T + + + ++ G K
Sbjct: 1195KPA---ETYFYDFIDLSGNEISGSAV-GLVNSTEYLVGFWGSGNKLKFDFERLKFGERFK 1362
Query: 409 YLSAFHNRFTGPVPTSLKSCSSIERIRIEGNQIEGDI 445
YL HN G VP KS + +E++ + N + G+I
Sbjct: 1363YLDLSHNLVFGKVP---KSVAGLEKLNVSYNHLCGEI 1464
Score = 33.9 bits (76), Expect = 0.14
Identities = 29/78 (37%), Positives = 39/78 (49%), Gaps = 11/78 (14%)
Frame = +1
Query: 595 DSALASIDLSGNRLN------GNIPTSLGFLVQLS---MLNLSHNMLSGTIPS-TFSM-S 643
D + S+ LSGN N G I SL + L +LNL + +SG P F +
Sbjct: 319 DKRVTSLYLSGNPENPKSFFSGTISPSLSKIKNLDGFYLLNLKN--ISGPFPGFLFKLPK 492
Query: 644 LDFVNISDNQLDGPLPEN 661
L F+ I +NQL G +PEN
Sbjct: 493 LQFIYIENNQLSGRIPEN 546
>TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, partial (64%)
Length = 969
Score = 156 bits (394), Expect = 2e-38
Identities = 83/217 (38%), Positives = 125/217 (57%), Gaps = 2/217 (0%)
Frame = +2
Query: 759 FENIIEATENFDDKYLIGVGSQGNVYKAELPTGLVVAVKKLHLVRDEEMSFFSSKSFTSE 818
++ + AT F D +G G G+VY GL +AVKKL + M+ + F E
Sbjct: 272 YKELHAATGGFSDDNKLGEGGFGSVYWGRTSDGLQIAVKKL-----KAMNSKAEMEFAVE 436
Query: 819 IETLTGIKHRNIIKLHGFCSHSKFSFLVYKFMEGGSLDQILNNEKQA-IAFDWEKRVNVV 877
+E L ++H+N++ L G+C +VY +M SL L+ + + +W+KR+ +
Sbjct: 437 VEVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQFAVEVQLNWQKRMKIA 616
Query: 878 KGVANALSYLHHDCSPPIIHRDISSKNILLNLDYEAHVSDFGTAKFLKPDL-HSWTQFAG 936
G A + YLHH+ +P IIHRDI + N+LLN D+E V+DFG AK + + H T+ G
Sbjct: 617 IGSAEGILYLHHEVTPHIIHRDIKASNVLLNSDFEPLVADFGFAKLIPEGVSHMTTRVKG 796
Query: 937 TFGYAAPELSQTMEVNEKCDVYSFGVLALEIIIGKHP 973
T GY APE + +V+E CDVYSFG+L LE++ G+ P
Sbjct: 797 TLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKP 907
>TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partial (77%)
Length = 1478
Score = 152 bits (385), Expect = 2e-37
Identities = 92/242 (38%), Positives = 136/242 (56%), Gaps = 10/242 (4%)
Frame = +1
Query: 744 KGVLFSIWSHDGKMM-----FENIIEATENFDDKYLIGVGSQGNVYKAELPTGLVVAVKK 798
KGV S S +GK F + +AT NF + LIG G G VYK L TG VAVK+
Sbjct: 361 KGVSSSNGSSNGKTAAASFGFRELADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQ 540
Query: 799 LHLVRDEEMSFFSSKSFTSEIETLTGIKHRNIIKLHGFCSHSKFSFLVYKFMEGGSLDQI 858
L D F + F E+ L+ + H N+++L G+C+ LVY++M GSL+
Sbjct: 541 LS--HDGRQGF---QEFVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDH 705
Query: 859 L---NNEKQAIAFDWEKRVNVVKGVANALSYLHHDCSPPIIHRDISSKNILLNLDYEAHV 915
L +++K+ + +W R+ V G A L YLH PP+I+RD+ S NILL+ ++ +
Sbjct: 706 LFELSHDKEPL--NWSTRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKL 879
Query: 916 SDFGTAKF--LKPDLHSWTQFAGTFGYAAPELSQTMEVNEKCDVYSFGVLALEIIIGKHP 973
SDFG AK + + H T+ GT+GY APE + + ++ K D+YSFGV+ LE++ G+
Sbjct: 880 SDFGLAKLGPVGDNTHVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRA 1059
Query: 974 GD 975
D
Sbjct: 1060ID 1065
>BP033681
Length = 454
Score = 151 bits (382), Expect = 5e-37
Identities = 75/124 (60%), Positives = 98/124 (78%)
Frame = +1
Query: 6 FIIMILCVLPTLSVAEDSEAKLALLKWKASFDNQSQSILSTWKNTTNPCSKWRGIECDKS 65
F+ ++L +L + AE+SEA ALLKWK S DNQSQ++LSTW + +PC KWRGI+CDKS
Sbjct: 91 FLYILLTLLLPQAAAEESEAN-ALLKWKQSLDNQSQALLSTWTGS-DPC-KWRGIQCDKS 261
Query: 66 NLISTIDLANLGLKGTLHSLTFSSFPNLITLNIYNNHFYGTIPPQIGNLSRINTLNFSKN 125
+ IS I LAN GLKGTLH+L+FSSFPNL T++I+ N FYGTIPPQ+G++ ++N LNFS+N
Sbjct: 262 SSISRISLANYGLKGTLHTLSFSSFPNLKTIDIFGNSFYGTIPPQVGDMPKVNILNFSEN 441
Query: 126 PIIG 129
G
Sbjct: 442 SFDG 453
Score = 40.0 bits (92), Expect = 0.002
Identities = 16/37 (43%), Positives = 25/37 (67%)
Frame = +1
Query: 552 LEELDLGGNELSGTIPNEVAELPKLRMLNLSRNRIEG 588
L+ +D+ GN GTIP +V ++PK+ +LN S N +G
Sbjct: 343 LKTIDIFGNSFYGTIPPQVGDMPKVNILNFSENSFDG 453
Score = 34.7 bits (78), Expect = 0.081
Identities = 19/64 (29%), Positives = 30/64 (46%), Gaps = 1/64 (1%)
Frame = +1
Query: 429 SSIERIRIEGNQIEGDIAE-DFGVYPNLRYVDLSDNKFHGHISPNWGKSLDLETFMISNT 487
SSI RI + ++G + F +PNL+ +D+ N F+G I P G + S
Sbjct: 262 SSISRISLANYGLKGTLHTLSFSSFPNLKTIDIFGNSFYGTIPPQVGDMPKVNILNFSEN 441
Query: 488 NISG 491
+ G
Sbjct: 442 SFDG 453
Score = 29.6 bits (65), Expect = 2.6
Identities = 16/63 (25%), Positives = 32/63 (50%), Gaps = 1/63 (1%)
Frame = +1
Query: 310 NLTLLLLRNNRLSGSIPA-SIGNLINLKYFSVQVNNLTGTIPATIGNLKQLIVFEVASNK 368
+++ + L N L G++ S + NLK + N+ GTIP +G++ ++ + + N
Sbjct: 265 SISRISLANYGLKGTLHTLSFSSFPNLKTIDIFGNSFYGTIPPQVGDMPKVNILNFSENS 444
Query: 369 LYG 371
G
Sbjct: 445 FDG 453
>TC12524 similar to UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1,
partial (27%)
Length = 834
Score = 148 bits (374), Expect = 4e-36
Identities = 100/302 (33%), Positives = 148/302 (48%), Gaps = 1/302 (0%)
Frame = +2
Query: 216 YIDLSNNFLSGVIPETIGNMSKLNQLMF-ANNTKLYGPIPHSLWNMSSLTLIYLYNMSLS 274
Y+ NN+ G+ PE IGN+++L L F A L G IP L + L ++L LS
Sbjct: 8 YLGYYNNYQGGIPPE-IGNLTQL--LRFDAAYCGLSGEIPAELGKLQKLDTLFLQVNVLS 178
Query: 275 GSIPDSVQNLINLDVLALYMNNLSGFIPSTIGNLKNLTLLLLRNNRLSGSIPASIGNLIN 334
GS+ + +L +L + L N LSG +P++ LKNLTLL L NRL G+IP +G +
Sbjct: 179 GSLTPELGHLKSLKSMDLSNNMLSGQVPASFAELKNLTLLNLFRNRLHGAIPEFVGEMPA 358
Query: 335 LKYFSVQVNNLTGTIPATIGNLKQLIVFEVASNKLYGRIPNGLYNITNWYSFVVSENDFV 394
L+ + NN TG+IP ++G +L + +++SNKL
Sbjct: 359 LEVLQLWENNFTGSIPQSLGKNGKLTLVDLSSNKL------------------------T 466
Query: 395 GHLPSQMCTGGSLKYLSAFHNRFTGPVPTSLKSCSSIERIRIEGNQIEGDIAEDFGVYPN 454
G LP MC+G L+ L A N GP+P SL C S+ RIR+ N + G I + P
Sbjct: 467 GTLPPHMCSGNRLQTLIALGNFLFGPIPESLGKCESLTRIRMGQNFLNGSIPKGLFGLPK 646
Query: 455 LRYVDLSDNKFHGHISPNWGKSLDLETFMISNTNISGGIPLDFIGLTKLGRLHLSSNQLT 514
L V+ DN G S ++ +SN +SG +P T + +L L N+ +
Sbjct: 647 LTQVEFQDNLLSGEFPETGSVSHNIGQITLSNNKLSGPLPSTIGNFTSMQKLLLDGNKFS 826
Query: 515 GK 516
G+
Sbjct: 827 GR 832
Score = 147 bits (372), Expect = 7e-36
Identities = 86/271 (31%), Positives = 145/271 (52%)
Frame = +2
Query: 173 NNFSGGPIPPEIGKLKKLRYLAITQGSLVGSIPQEIGLLTNLTYIDLSNNFLSGVIPETI 232
NN+ GG IPPEIG L +L L G IP E+G L L + L N LSG + +
Sbjct: 23 NNYQGG-IPPEIGNLTQLLRFDAAYCGLSGEIPAELGKLQKLDTLFLQVNVLSGSLTPEL 199
Query: 233 GNMSKLNQLMFANNTKLYGPIPHSLWNMSSLTLIYLYNMSLSGSIPDSVQNLINLDVLAL 292
G++ L + +NN L G +P S + +LTL+ L+ L G+IP+ V + L+VL L
Sbjct: 200 GHLKSLKSMDLSNN-MLSGQVPASFAELKNLTLLNLFRNRLHGAIPEFVGEMPALEVLQL 376
Query: 293 YMNNLSGFIPSTIGNLKNLTLLLLRNNRLSGSIPASIGNLINLKYFSVQVNNLTGTIPAT 352
+ NN +G IP ++G LTL+ L +N+L+G++P + + L+ N L G IP +
Sbjct: 377 WENNFTGSIPQSLGKNGKLTLVDLSSNKLTGTLPPHMCSGNRLQTLIALGNFLFGPIPES 556
Query: 353 IGNLKQLIVFEVASNKLYGRIPNGLYNITNWYSFVVSENDFVGHLPSQMCTGGSLKYLSA 412
+G + L + N L G IP GL+ + +N G P ++ ++
Sbjct: 557 LGKCESLTRIRMGQNFLNGSIPKGLFGLPKLTQVEFQDNLLSGEFPETGSVSHNIGQITL 736
Query: 413 FHNRFTGPVPTSLKSCSSIERIRIEGNQIEG 443
+N+ +GP+P+++ + +S++++ ++GN+ G
Sbjct: 737 SNNKLSGPLPSTIGNFTSMQKLLLDGNKFSG 829
Score = 140 bits (352), Expect = 1e-33
Identities = 92/273 (33%), Positives = 140/273 (50%), Gaps = 24/273 (8%)
Frame = +2
Query: 99 YNNHFYGTIPPQIGNLS------------------------RINTLNFSKNPIIGSIPQE 134
Y N++ G IPP+IGNL+ +++TL N + GS+ E
Sbjct: 17 YYNNYQGGIPPEIGNLTQLLRFDAAYCGLSGEIPAELGKLQKLDTLFLQVNVLSGSLTPE 196
Query: 135 MYTLRSLKGLDFFFCTLSGEIDKSIGNLTNLSYLDLGGNNFSGGPIPPEIGKLKKLRYLA 194
+ L+SLK +D LSG++ S L NL+ L+L N G IP +G++ L L
Sbjct: 197 LGHLKSLKSMDLSNNMLSGQVPASFAELKNLTLLNLFRNRLHGA-IPEFVGEMPALEVLQ 373
Query: 195 ITQGSLVGSIPQEIGLLTNLTYIDLSNNFLSGVIPETIGNMSKLNQLMFANNTKLYGPIP 254
+ + + GSIPQ +G LT +DLS+N L+G +P + + ++L L+ N L+GPIP
Sbjct: 374 LWENNFTGSIPQSLGKNGKLTLVDLSSNKLTGTLPPHMCSGNRLQTLIALGNF-LFGPIP 550
Query: 255 HSLWNMSSLTLIYLYNMSLSGSIPDSVQNLINLDVLALYMNNLSGFIPSTIGNLKNLTLL 314
SL SLT I + L+GSIP + L L + N LSG P T N+ +
Sbjct: 551 ESLGKCESLTRIRMGQNFLNGSIPKGLFGLPKLTQVEFQDNLLSGEFPETGSVSHNIGQI 730
Query: 315 LLRNNRLSGSIPASIGNLINLKYFSVQVNNLTG 347
L NN+LSG +P++IGN +++ + N +G
Sbjct: 731 TLSNNKLSGPLPSTIGNFTSMQKLLLDGNKFSG 829
Score = 124 bits (312), Expect = 6e-29
Identities = 80/251 (31%), Positives = 131/251 (51%), Gaps = 4/251 (1%)
Frame = +2
Query: 413 FHNRFTGPVPTSLKSCSSIERIRIEGNQIEGDIAEDFGVYPNLRYVDLSDNKFHGHISPN 472
++N + G +P + + + + R + G+I + G L + L N G ++P
Sbjct: 17 YYNNYQGGIPPEIGNLTQLLRFDAAYCGLSGEIPAELGKLQKLDTLFLQVNVLSGSLTPE 196
Query: 473 WGKSLDLETFMISNTNISGGIPLDFIGLTKLGRLHLSSNQLTGKLPKEILGGMKSLLYLK 532
G L++ +SN +SG +P F L L L+L N+L G +P E +G M +L L+
Sbjct: 197 LGHLKSLKSMDLSNNMLSGQVPASFAELKNLTLLNLFRNRLHGAIP-EFVGEMPALEVLQ 373
Query: 533 ISNNHFTDSIPTEIGLLQRLEELDLGGNELSGTIPNEVAELPKLRMLNLSRNRIEGRIPS 592
+ N+FT SIP +G +L +DL N+L+GT+P + +L+ L N + G IP
Sbjct: 374 LWENNFTGSIPQSLGKNGKLTLVDLSSNKLTGTLPPHMCSGNRLQTLIALGNFLFGPIPE 553
Query: 593 TFDS--ALASIDLSGNRLNGNIPTSLGFLVQLSMLNLSHNMLSGTIPSTFSMS--LDFVN 648
+ +L I + N LNG+IP L L +L+ + N+LSG P T S+S + +
Sbjct: 554 SLGKCESLTRIRMGQNFLNGSIPKGLFGLPKLTQVEFQDNLLSGEFPETGSVSHNIGQIT 733
Query: 649 ISDNQLDGPLP 659
+S+N+L GPLP
Sbjct: 734 LSNNKLSGPLP 766
Score = 118 bits (296), Expect = 4e-27
Identities = 75/271 (27%), Positives = 122/271 (44%)
Frame = +2
Query: 319 NRLSGSIPASIGNLINLKYFSVQVNNLTGTIPATIGNLKQLIVFEVASNKLYGRIPNGLY 378
N G IP IGNL L F L+G IPA +G L++L + N L G + L
Sbjct: 23 NNYQGGIPPEIGNLTQLLRFDAAYCGLSGEIPAELGKLQKLDTLFLQVNVLSGSLTPELG 202
Query: 379 NITNWYSFVVSENDFVGHLPSQMCTGGSLKYLSAFHNRFTGPVPTSLKSCSSIERIRIEG 438
++ + S +S N G +P+ +L L+ F NR G +P + ++E +++
Sbjct: 203 HLKSLKSMDLSNNMLSGQVPASFAELKNLTLLNLFRNRLHGAIPEFVGEMPALEVLQLWE 382
Query: 439 NQIEGDIAEDFGVYPNLRYVDLSDNKFHGHISPNWGKSLDLETFMISNTNISGGIPLDFI 498
N G I + G L VDLS NK G + P+ L+T + + G IP
Sbjct: 383 NNFTGSIPQSLGKNGKLTLVDLSSNKLTGTLPPHMCSGNRLQTLIALGNFLFGPIPESLG 562
Query: 499 GLTKLGRLHLSSNQLTGKLPKEILGGMKSLLYLKISNNHFTDSIPTEIGLLQRLEELDLG 558
L R+ + N L G +PK + G+ L ++ +N + P + + ++ L
Sbjct: 563 KCESLTRIRMGQNFLNGSIPKGLF-GLPKLTQVEFQDNLLSGEFPETGSVSHNIGQITLS 739
Query: 559 GNELSGTIPNEVAELPKLRMLNLSRNRIEGR 589
N+LSG +P+ + ++ L L N+ GR
Sbjct: 740 NNKLSGPLPSTIGNFTSMQKLLLDGNKFSGR 832
Score = 114 bits (286), Expect = 6e-26
Identities = 80/270 (29%), Positives = 128/270 (46%), Gaps = 2/270 (0%)
Frame = +2
Query: 343 NNLTGTIPATIGNLKQLIVFEVASNKLYGRIPNGLYNITNWYSFVVSENDFVGHLPSQMC 402
NN G IP IGNL QL+ F+ A L G IP L + + + N G L ++
Sbjct: 23 NNYQGGIPPEIGNLTQLLRFDAAYCGLSGEIPAELGKLQKLDTLFLQVNVLSGSLTPELG 202
Query: 403 TGGSLKYLSAFHNRFTGPVPTSLKSCSSIERIRIEGNQIEGDIAEDFGVYPNLRYVDLSD 462
SLK + +N +G VP S ++ + + N++ G I E G P L + L +
Sbjct: 203 HLKSLKSMDLSNNMLSGQVPASFAELKNLTLLNLFRNRLHGAIPEFVGEMPALEVLQLWE 382
Query: 463 NKFHGHISPNWGKSLDLETFMISNTNISGGIPLDFIGLTKLGRLHLSSNQLTGKLPKEIL 522
N F G I + GK+ L +S+ ++G +P +L L N L G +P E L
Sbjct: 383 NNFTGSIPQSLGKNGKLTLVDLSSNKLTGTLPPHMCSGNRLQTLIALGNFLFGPIP-ESL 559
Query: 523 GGMKSLLYLKISNNHFTDSIPTEIGLLQRLEELDLGGNELSGTIPNEVAELPKLRMLNLS 582
G +SL +++ N SIP + L +L +++ N LSG P + + + LS
Sbjct: 560 GKCESLTRIRMGQNFLNGSIPKGLFGLPKLTQVEFQDNLLSGEFPETGSVSHNIGQITLS 739
Query: 583 RNRIEGRIPSTFD--SALASIDLSGNRLNG 610
N++ G +PST +++ + L GN+ +G
Sbjct: 740 NNKLSGPLPSTIGNFTSMQKLLLDGNKFSG 829
Score = 101 bits (252), Expect = 5e-22
Identities = 69/186 (37%), Positives = 101/186 (54%), Gaps = 4/186 (2%)
Frame = +2
Query: 480 ETFMISNTNISGGIPLDFIGLTKLGRLHLSSNQLTGKLPKEILGGMKSLLYLKISNNHFT 539
E ++ N GGIP + LT+L R + L+G++P E LG ++ L L + N +
Sbjct: 2 ELYLGYYNNYQGGIPPEIGNLTQLLRFDAAYCGLSGEIPAE-LGKLQKLDTLFLQVNVLS 178
Query: 540 DSIPTEIGLLQRLEELDLGGNELSGTIPNEVAELPKLRMLNLSRNRIEGRIPSTFDS--A 597
S+ E+G L+ L+ +DL N LSG +P AEL L +LNL RNR+ G IP A
Sbjct: 179 GSLTPELGHLKSLKSMDLSNNMLSGQVPASFAELKNLTLLNLFRNRLHGAIPEFVGEMPA 358
Query: 598 LASIDLSGNRLNGNIPTSLGFLVQLSMLNLSHNMLSGTIPSTFSMS--LDFVNISDNQLD 655
L + L N G+IP SLG +L++++LS N L+GT+P L + N L
Sbjct: 359 LEVLQLWENNFTGSIPQSLGKNGKLTLVDLSSNKLTGTLPPHMCSGNRLQTLIALGNFLF 538
Query: 656 GPLPEN 661
GP+PE+
Sbjct: 539 GPIPES 556
Score = 57.4 bits (137), Expect = 1e-08
Identities = 44/163 (26%), Positives = 74/163 (44%), Gaps = 3/163 (1%)
Frame = +2
Query: 43 ILSTWKNT-TNPCSKWRGIECDKSNLISTIDLANLGLKGTL--HSLTFSSFPNLITLNIY 99
+L W+N T + G K+ ++ +DL++ L GTL H + + LI L
Sbjct: 365 VLQLWENNFTGSIPQSLG----KNGKLTLVDLSSNKLTGTLPPHMCSGNRLQTLIALG-- 526
Query: 100 NNHFYGTIPPQIGNLSRINTLNFSKNPIIGSIPQEMYTLRSLKGLDFFFCTLSGEIDKSI 159
N +G IP +G + + +N + GSIP+ ++ L L ++F LSGE ++
Sbjct: 527 -NFLFGPIPESLGKCESLTRIRMGQNFLNGSIPKGLFGLPKLTQVEFQDNLLSGEFPETG 703
Query: 160 GNLTNLSYLDLGGNNFSGGPIPPEIGKLKKLRYLAITQGSLVG 202
N+ + L N S GP+P IG ++ L + G
Sbjct: 704 SVSHNIGQITLSNNKLS-GPLPSTIGNFTSMQKLLLDGNKFSG 829
>TC17311 similar to UP|Q8K2Y2 (Q8K2Y2) Receptor-interacting serine-threonine
kinase 3, partial (7%)
Length = 606
Score = 143 bits (360), Expect = 2e-34
Identities = 75/165 (45%), Positives = 104/165 (62%), Gaps = 11/165 (6%)
Frame = +3
Query: 818 EIETLTGIKHRNIIKLHGFCSHSKFSFLVYKFMEGGSLDQILNNEK---------QAIAF 868
E+E L+ I+H NI+KL S + LVY ++E SLD+ L+ + Q
Sbjct: 3 EVEILSNIRHNNIVKLQCCISSEESLLLVYDYLENLSLDRWLHKKSKPPTVSGSVQNNII 182
Query: 869 DWEKRVNVVKGVANALSYLHHDCSPPIIHRDISSKNILLNLDYEAHVSDFGTAKF-LKP- 926
DW +R+++ GVA L Y+HHDCSPP++HRD+ NILL+ + A V+DFG A +KP
Sbjct: 183 DWPRRLHIAIGVAQGLCYMHHDCSPPVVHRDLKPSNILLDSQFNAKVADFGLAMMSVKPE 362
Query: 927 DLHSWTQFAGTFGYAAPELSQTMEVNEKCDVYSFGVLALEIIIGK 971
+L + + AGTFGY APE + T+ VNEK DVYSFGV+ LE+ GK
Sbjct: 363 ELATMSAVAGTFGYIAPEYALTIRVNEKIDVYSFGVILLELTTGK 497
>CN825263
Length = 663
Score = 142 bits (357), Expect = 4e-34
Identities = 81/199 (40%), Positives = 114/199 (56%), Gaps = 3/199 (1%)
Frame = +1
Query: 781 GNVYKAELPTGLVVAVKKLHLVRDEEMSFFSSKSFTSEIETLTGIKHRNIIKLHGFCSHS 840
G VYK L G VAVK L RD++ + F +E+E L+ + HRN++KL G C
Sbjct: 7 GLVYKGILNDGRDVAVKILK--RDDQRG---GREFLAEVEMLSRLHHRNLVKLIGICIEK 171
Query: 841 KFSFLVYKFMEGGSLDQILNN-EKQAIAFDWEKRVNVVKGVANALSYLHHDCSPPIIHRD 899
+ L+Y+ + GS++ L+ +K+ DW R+ + G A L+YLH D +P +IHRD
Sbjct: 172 QTRCLIYELVPNGSVESHLHGADKETGPLDWNARMKIALGAARGLAYLHEDSNPCVIHRD 351
Query: 900 ISSKNILLNLDYEAHVSDFGTAKFL--KPDLHSWTQFAGTFGYAAPELSQTMEVNEKCDV 957
S NILL D+ VSDFG A+ + + H T GTFGY APE + T + K DV
Sbjct: 352 FKSSNILLECDFTPKVSDFGLARTALDEGNKHISTHVMGTFGYLAPEYAMTGHLLVKSDV 531
Query: 958 YSFGVLALEIIIGKHPGDL 976
YS+GV+ LE++ G P DL
Sbjct: 532 YSYGVVLLELLTGTKPVDL 588
>TC15149 similar to UP|Q9LVI6 (Q9LVI6) Probable receptor-like protein kinase
protein (AT3g17840/MEB5_6), partial (45%)
Length = 1489
Score = 141 bits (355), Expect = 6e-34
Identities = 93/274 (33%), Positives = 141/274 (50%), Gaps = 6/274 (2%)
Frame = +3
Query: 774 LIGVGSQGNVYKAELPTGLVVAVKKLHLVRDEEMSFFSSKSFTSEIETLTGIKHRNIIKL 833
++G G+ G YKA L TGLVVAVK+L V S K F +IET+ + H++++ L
Sbjct: 291 VLGKGTFGTAYKAVLETGLVVAVKRLKDVT------ISEKEFKDKIETVGAMDHQSLVPL 452
Query: 834 HGFCSHSKFSFLVYKFMEGGSLDQILNNEKQA--IAFDWEKRVNVVKGVANALSYLHHDC 891
+ LVY +M GSL +L+ K A +WE R + G A + YLH
Sbjct: 453 RAYYFSRDEKLLVYDYMPMGSLSALLHGNKGAGRTPLNWEIRSGIALGAARGIEYLHSQ- 629
Query: 892 SPPIIHRDISSKNILLNLDYEAHVSDFGTAKFLKPDLHSWTQFAGTFGYAAPELSQTMEV 951
P + H +I + NILL YEA VSDFG A + P + GY APE++ +V
Sbjct: 630 GPNVSHGNIKASNILLTKSYEAKVSDFGLAHLVGPS----STPNRVAGYRAPEVTDPRKV 797
Query: 952 NEKCDVYSFGVLALEIIIGKHPGDLI----SLFLSPSTRPTANDMLLTEVLDQRPQKVIK 1007
++K DVYSFGVL LE++ GK P + + L + + +EV D + +
Sbjct: 798 SQKADVYSFGVLLLELLTGKAPTHALLNEEGVDLPRWVQSLVREEWTSEVFDLELLR-YQ 974
Query: 1008 PIDEEVILIAKLAFSCLNQVPRSRPTMDQVCKML 1041
++EE++ + +LA C P RP++ +V + +
Sbjct: 975 NVEEEMVQLLQLAVDCAAPYPDRRPSIAEVTRSI 1076
>TC9538 similar to UP|RLK5_ARATH (P47735) Receptor-like protein kinase 5
precursor , partial (10%)
Length = 1067
Score = 141 bits (355), Expect = 6e-34
Identities = 87/232 (37%), Positives = 127/232 (54%), Gaps = 19/232 (8%)
Frame = +2
Query: 829 NIIKLHGFCSHSKFSFLVYKFMEGGSLDQILNNE----------KQAIAFDWEKRVNVVK 878
NI++L S+ LVY+++E SLD+ L+ + +Q DW KR+ +
Sbjct: 5 NIVRLLCCISNEASMLLVYEYLENHSLDKWLHLKPKSSSVSGVVQQYTVLDWPKRLKIAI 184
Query: 879 GVANALSYLHHDCSPPIIHRDISSKNILLNLDYEAHVSDFGTAKFL-KP-DLHSWTQFAG 936
G A LSY+HHDCSPPI+HRD+ + NILL+ + A V+DFG A+ L KP +L+ + G
Sbjct: 185 GAAQGLSYMHHDCSPPIVHRDVKTSNILLDKQFNAKVADFGLARMLIKPGELNIMSTVIG 364
Query: 937 TFGYAAPELSQTMEVNEKCDVYSFGVLALEIIIGKHP--GDLISLFLSPSTR-----PTA 989
TFGY APE QT ++EK DVYSFGV+ LE+ GK GD S + R
Sbjct: 365 TFGYIAPEYVQTTRISEKVDVYSFGVVLLELTTGKEANYGDQHSSLAEWAWRHILIGSNV 544
Query: 990 NDMLLTEVLDQRPQKVIKPIDEEVILIAKLAFSCLNQVPRSRPTMDQVCKML 1041
D+L +V++ +E+ + KL C +P +RP+M +V +L
Sbjct: 545 XDLLXKDVME-------ASYIDEMCSVFKLGVMCTATLPATRPSMKEVLPIL 679
>AV416024
Length = 249
Score = 139 bits (351), Expect = 2e-33
Identities = 68/82 (82%), Positives = 73/82 (88%)
Frame = +2
Query: 769 FDDKYLIGVGSQGNVYKAELPTGLVVAVKKLHLVRDEEMSFFSSKSFTSEIETLTGIKHR 828
FDDKYLIGVG QGNVY AELP GLVVAVKKLH V DEEMS FSSK+F SEI+TLT I+HR
Sbjct: 2 FDDKYLIGVGGQGNVYMAELPAGLVVAVKKLHSVTDEEMSNFSSKAFNSEIQTLTEIRHR 181
Query: 829 NIIKLHGFCSHSKFSFLVYKFM 850
NIIKLHGFC HSKFSFLVY+F+
Sbjct: 182 NIIKLHGFCLHSKFSFLVYEFL 247
>TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2, complete
Length = 2946
Score = 137 bits (344), Expect = 1e-32
Identities = 106/384 (27%), Positives = 178/384 (45%), Gaps = 23/384 (5%)
Frame = +1
Query: 685 LVPCATSQIHSRKSKNI--LQSVFIALGALILVLSGVGISMYVFFRRKKPN--------- 733
+V + ++KS N L + + A ++ ++ +G+++ +RKK
Sbjct: 1402 VVASKLDRTRNKKSINTKKLAGSLVVIIAFVIFITILGLAISTCIQRKKNKRGDEGEIGI 1581
Query: 734 ----EEIQTEEEVQKGVLFSIWSHDGKMMFENIIEATENFDDKYLIGVGSQGNVYKAELP 789
++ + +E++ +F F I AT +F +G G G VYK L
Sbjct: 1582 INHWKDKRGDEDIDLATIFD---------FSTISSATNHFSLSNKLGEGGFGPVYKGLLA 1734
Query: 790 TGLVVAVKKLHLVRDEEMSFFSSKSFTSEIETLTGIKHRNIIKLHGFCSHSKFSFLVYKF 849
G +AVK+L + M + F +EI+ + ++HRN++KL G H + K
Sbjct: 1735 NGQEIAVKRLSNTSGQGM-----EEFKNEIKLIARLQHRNLVKLFGCSVHQDENSHANKK 1899
Query: 850 MEGGSLDQILNNEKQAIAFDWEKRVNVVKGVANALSYLHHDCSPPIIHRDISSKNILLNL 909
M+ IL + ++ DW KR+ ++ G+A L YLH D IIHRD+ + NILL+
Sbjct: 1900 MK------ILLDSTRSKLVDWNKRLQIIDGIARGLLYLHQDSRLRIIHRDLKTSNILLDD 2061
Query: 910 DYEAHVSDFGTAK-FLKPDLHSWT-QFAGTFGYAAPELSQTMEVNEKCDVYSFGVLALEI 967
+ +SDFG A+ F+ + + T + GT+GY PE + + K DV+SFGV+ LEI
Sbjct: 2062 EMNPKISDFGLARIFIGDQVEARTKRVMGTYGYMPPEYAVHGSFSIKSDVFSFGVIVLEI 2241
Query: 968 IIGKHPGDLISLFLSPSTRPTANDMLLTEVLDQRPQKVIKP------IDEEVILIAKLAF 1021
I GK I F P +++RP +++ I E++ +A
Sbjct: 2242 ISGKK----IGRFYDPHHHLNLLSHAWRLWIEERPLELVDELLDDPVIPTEILRYIHVAL 2409
Query: 1022 SCLNQVPRSRPTMDQVCKMLGAGK 1045
C+ + P +RP M + ML K
Sbjct: 2410 LCVQRRPENRPDMLSIVLMLNGEK 2481
>TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete
Length = 2193
Score = 135 bits (339), Expect = 4e-32
Identities = 95/288 (32%), Positives = 144/288 (49%), Gaps = 12/288 (4%)
Frame = +1
Query: 759 FENIIEATENFDDKYLIGVGSQGNVYKAELPTGLVVAVKKLHLVRDEEMSFFSSKSFTSE 818
++ + +AT NF IG G G VY AEL G A+KK M +S F E
Sbjct: 1057 YQELAKATNNFSLDNKIGQGGFGAVYYAEL-RGKKTAIKK--------MDVQASTEFLCE 1209
Query: 819 IETLTGIKHRNIIKLHGFCSHSKFSFLVYKFMEGGSLDQILNNEKQAIAFDWEKRVNVVK 878
++ LT + H N+++L G+C FLVY+ ++ G+L Q L+ + W RV +
Sbjct: 1210 LKVLTHVHHLNLVRLIGYCVEGSL-FLVYEHIDNGNLGQYLHGSGKE-PLPWSSRVQIAL 1383
Query: 879 GVANALSYLHHDCSPPIIHRDISSKNILLNLDYEAHVSDFGTAKFLKPDLHS-WTQFAGT 937
A L Y+H P IHRD+ S NIL++ + V+DFG K ++ + T+ GT
Sbjct: 1384 DAARGLEYIHEHTVPVYIHRDVKSANILIDKNLRGKVADFGLTKLIEVGNSTLQTRLVGT 1563
Query: 938 FGYAAPELSQTMEVNEKCDVYSFGVLALEIIIGKH-----------PGDLISLFLSPSTR 986
FGY PE +Q +++ K DVY+FGV+ E+I K+ L++LF +
Sbjct: 1564 FGYMPPEYAQYGDISPKIDVYAFGVVLFELISAKNAVLKTGELVAESKGLVALFEEALNK 1743
Query: 987 PTANDMLLTEVLDQRPQKVIKPIDEEVILIAKLAFSCLNQVPRSRPTM 1034
D L +++D R + PID V+ IA+L +C P RP+M
Sbjct: 1744 SDPCD-ALRKLVDPRLGENY-PID-SVLKIAQLGRACTRDNPLLRPSM 1878
>AW719637
Length = 571
Score = 134 bits (338), Expect = 6e-32
Identities = 76/188 (40%), Positives = 107/188 (56%)
Frame = +2
Query: 178 GPIPPEIGKLKKLRYLAITQGSLVGSIPQEIGLLTNLTYIDLSNNFLSGVIPETIGNMSK 237
G IP EI L L+ L++ L G+IP IG L++L + L +N LSG IP++IG++
Sbjct: 8 GEIPEEICSLSNLQSLSLHTNFLEGNIPSNIGNLSSLVNLTLFDNHLSGEIPKSIGSLRN 187
Query: 238 LNQLMFANNTKLYGPIPHSLWNMSSLTLIYLYNMSLSGSIPDSVQNLINLDVLALYMNNL 297
L N L G IP + N +SL ++ L S+SGS+P S+Q L + +A+Y L
Sbjct: 188 LQVFRAGGNKNLKGEIPWEIGNCTSLVMLGLAETSISGSLPSSIQLLKRIKTIAIYTTLL 367
Query: 298 SGFIPSTIGNLKNLTLLLLRNNRLSGSIPASIGNLINLKYFSVQVNNLTGTIPATIGNLK 357
SG IP IGN L L L N +SGSIP+ IG L LK + NN+ GTIP IG
Sbjct: 368 SGSIPEEIGNCSELQNLYLYQNSISGSIPSQIGELSKLKSLLLWQNNIVGTIPEEIGRCT 547
Query: 358 QLIVFEVA 365
++ V +++
Sbjct: 548 EMEVIDLS 571
Score = 121 bits (303), Expect = 7e-28
Identities = 79/210 (37%), Positives = 102/210 (47%)
Frame = +2
Query: 105 GTIPPQIGNLSRINTLNFSKNPIIGSIPQEMYTLRSLKGLDFFFCTLSGEIDKSIGNLTN 164
G IP +I +LS + +L+ N + G+IP + L SL L F LSGEI KSIG+L N
Sbjct: 8 GEIPEEICSLSNLQSLSLHTNFLEGNIPSNIGNLSSLVNLTLFDNHLSGEIPKSIGSLRN 187
Query: 165 LSYLDLGGNNFSGGPIPPEIGKLKKLRYLAITQGSLVGSIPQEIGLLTNLTYIDLSNNFL 224
L GGN G IP EIG L L + + S+ GS+P I LL + I + L
Sbjct: 188 LQVFRAGGNKNLKGEIPWEIGNCTSLVMLGLAETSISGSLPSSIQLLKRIKTIAIYTTLL 367
Query: 225 SGVIPETIGNMSKLNQLMFANNTKLYGPIPHSLWNMSSLTLIYLYNMSLSGSIPDSVQNL 284
SG IPE IGN S+L L YLY S+SGSIP + L
Sbjct: 368 SGSIPEEIGNCSELQNL-------------------------YLYQNSISGSIPSQIGEL 472
Query: 285 INLDVLALYMNNLSGFIPSTIGNLKNLTLL 314
L L L+ NN+ G IP IG + ++
Sbjct: 473 SKLKSLLLWQNNIVGTIPEEIGRCTEMEVI 562
Score = 108 bits (271), Expect = 3e-24
Identities = 68/183 (37%), Positives = 101/183 (55%), Gaps = 1/183 (0%)
Frame = +2
Query: 200 LVGSIPQEIGLLTNLTYIDLSNNFLSGVIPETIGNMSKLNQLMFANNTKLYGPIPHSLWN 259
L+G IP+EI L+NL + L NFL G IP IGN+S L L +N L G IP S+ +
Sbjct: 2 LLGEIPEEICSLSNLQSLSLHTNFLEGNIPSNIGNLSSLVNLTLFDN-HLSGEIPKSIGS 178
Query: 260 MSSLTLIYLY-NMSLSGSIPDSVQNLINLDVLALYMNNLSGFIPSTIGNLKNLTLLLLRN 318
+ +L + N +L G IP + N +L +L L ++SG +PS+I LK + + +
Sbjct: 179 LRNLQVFRAGGNKNLKGEIPWEIGNCTSLVMLGLAETSISGSLPSSIQLLKRIKTIAIYT 358
Query: 319 NRLSGSIPASIGNLINLKYFSVQVNNLTGTIPATIGNLKQLIVFEVASNKLYGRIPNGLY 378
LSGSIP IGN L+ + N+++G+IP+ IG L +L + N + G IP +
Sbjct: 359 TLLSGSIPEEIGNCSELQNLYLYQNSISGSIPSQIGELSKLKSLLLWQNNIVGTIPEEIG 538
Query: 379 NIT 381
T
Sbjct: 539 RCT 547
Score = 99.4 bits (246), Expect = 3e-21
Identities = 62/190 (32%), Positives = 97/190 (50%), Gaps = 1/190 (0%)
Frame = +2
Query: 297 LSGFIPSTIGNLKNLTLLLLRNNRLSGSIPASIGNLINLKYFSVQVNNLTGTIPATIGNL 356
L G IP I +L NL L L N L G+IP++IGNL +L ++ N+L+G IP +IG+L
Sbjct: 2 LLGEIPEEICSLSNLQSLSLHTNFLEGNIPSNIGNLSSLVNLTLFDNHLSGEIPKSIGSL 181
Query: 357 KQLIVFEVASNK-LYGRIPNGLYNITNWYSFVVSENDFVGHLPSQMCTGGSLKYLSAFHN 415
+ L VF NK L G IP + N T+ ++E G LPS + +K ++ +
Sbjct: 182 RNLQVFRAGGNKNLKGEIPWEIGNCTSLVMLGLAETSISGSLPSSIQLLKRIKTIAIYTT 361
Query: 416 RFTGPVPTSLKSCSSIERIRIEGNQIEGDIAEDFGVYPNLRYVDLSDNKFHGHISPNWGK 475
+G +P + +CS ++ + + N I G I G L+ + L N G I G+
Sbjct: 362 LLSGSIPEEIGNCSELQNLYLYQNSISGSIPSQIGELSKLKSLLLWQNNIVGTIPEEIGR 541
Query: 476 SLDLETFMIS 485
++E +S
Sbjct: 542 CTEMEVIDLS 571
Score = 91.3 bits (225), Expect = 7e-19
Identities = 62/189 (32%), Positives = 96/189 (49%), Gaps = 3/189 (1%)
Frame = +2
Query: 419 GPVPTSLKSCSSIERIRIEGNQIEGDIAEDFGVYPNLRYVDLSDNKFHGHISPNWGKSLD 478
G +P + S S+++ + + N +EG+I + G +L + L DN G I + G +
Sbjct: 8 GEIPEEICSLSNLQSLSLHTNFLEGNIPSNIGNLSSLVNLTLFDNHLSGEIPKSIGSLRN 187
Query: 479 LETFMIS-NTNISGGIPLDFIGLTKLGRLHLSSNQLTGKLPKEILGGMKSLLYLKISNNH 537
L+ F N N+ G IP + T L L L+ ++G LP I +K + + I
Sbjct: 188 LQVFRAGGNKNLKGEIPWEIGNCTSLVMLGLAETSISGSLPSSIQL-LKRIKTIAIYTTL 364
Query: 538 FTDSIPTEIGLLQRLEELDLGGNELSGTIPNEVAELPKLRMLNLSRNRIEGRIPSTFD-- 595
+ SIP EIG L+ L L N +SG+IP+++ EL KL+ L L +N I G IP
Sbjct: 365 LSGSIPEEIGNCSELQNLYLYQNSISGSIPSQIGELSKLKSLLLWQNNIVGTIPEEIGRC 544
Query: 596 SALASIDLS 604
+ + IDLS
Sbjct: 545 TEMEVIDLS 571
Score = 88.2 bits (217), Expect = 6e-18
Identities = 60/184 (32%), Positives = 91/184 (48%), Gaps = 25/184 (13%)
Frame = +2
Query: 62 CDKSNLISTIDLANLGLKGTLHSLTFSSFPNLITLNIYNNHFYGTIPPQIGNLSRINT-- 119
C SNL S + L L+G + S + +L+ L +++NH G IP IG+L +
Sbjct: 29 CSLSNLQS-LSLHTNFLEGNIPS-NIGNLSSLVNLTLFDNHLSGEIPKSIGSLRNLQVFR 202
Query: 120 -----------------------LNFSKNPIIGSIPQEMYTLRSLKGLDFFFCTLSGEID 156
L ++ I GS+P + L+ +K + + LSG I
Sbjct: 203 AGGNKNLKGEIPWEIGNCTSLVMLGLAETSISGSLPSSIQLLKRIKTIAIYTTLLSGSIP 382
Query: 157 KSIGNLTNLSYLDLGGNNFSGGPIPPEIGKLKKLRYLAITQGSLVGSIPQEIGLLTNLTY 216
+ IGN + L L L N+ SG IP +IG+L KL+ L + Q ++VG+IP+EIG T +
Sbjct: 383 EEIGNCSELQNLYLYQNSISGS-IPSQIGELSKLKSLLLWQNNIVGTIPEEIGRCTEMEV 559
Query: 217 IDLS 220
IDLS
Sbjct: 560 IDLS 571
Score = 57.0 bits (136), Expect = 2e-08
Identities = 38/110 (34%), Positives = 55/110 (49%), Gaps = 5/110 (4%)
Frame = +2
Query: 562 LSGTIPNEVAELPKLRMLNLSRNRIEGRIPSTFD--SALASIDLSGNRLNGNIPTSLGFL 619
L G IP E+ L L+ L+L N +EG IPS S+L ++ L N L+G IP S+G L
Sbjct: 2 LLGEIPEEICSLSNLQSLSLHTNFLEGNIPSNIGNLSSLVNLTLFDNHLSGEIPKSIGSL 181
Query: 620 VQLSMLNLSHNM-LSGTIPSTFS--MSLDFVNISDNQLDGPLPENPAFLR 666
L + N L G IP SL + +++ + G LP + L+
Sbjct: 182 RNLQVFRAGGNKNLKGEIPWEIGNCTSLVMLGLAETSISGSLPSSIQLLK 331
>TC19546 similar to UP|Q40543 (Q40543) Protein-serine/threonine kinase ,
partial (46%)
Length = 731
Score = 129 bits (324), Expect = 2e-30
Identities = 80/215 (37%), Positives = 116/215 (53%), Gaps = 11/215 (5%)
Frame = +1
Query: 740 EEVQKGVLFSIWSHDG-----------KMMFENIIEATENFDDKYLIGVGSQGNVYKAEL 788
E +QK FS WSH K +++I +AT+NF ++G GS G VYKA +
Sbjct: 115 ENLQKKSAFSWWSHQNNDQFASPSGIPKYSYKDIQKATQNFTT--ILGQGSFGTVYKATM 288
Query: 789 PTGLVVAVKKLHLVRDEEMSFFSSKSFTSEIETLTGIKHRNIIKLHGFCSHSKFSFLVYK 848
PTG VVAVK L S F +E+ L + HRN++ L GFC LVY+
Sbjct: 289 PTGEVVAVKVL-----APNSKQGEHEFQTEVHLLGRLHHRNLVNLVGFCVDKGQRILVYQ 453
Query: 849 FMEGGSLDQILNNEKQAIAFDWEKRVNVVKGVANALSYLHHDCSPPIIHRDISSKNILLN 908
FM GSL +L E++ ++ W++R+ + +++ + YLH PP+IHRD+ S NILL+
Sbjct: 454 FMSNGSLANLLYGEEKELS--WDERLQIAMDISHGIEYLHEGAVPPVIHRDLKSANILLD 627
Query: 909 LDYEAHVSDFGTAKFLKPDLHSWTQFAGTFGYAAP 943
A V+DFG +K D + + GT+GY P
Sbjct: 628 DSMRAMVADFGLSKEEIFDGRN-SGLKGTYGYMDP 729
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.138 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,001,996
Number of Sequences: 28460
Number of extensions: 250711
Number of successful extensions: 2647
Number of sequences better than 10.0: 375
Number of HSP's better than 10.0 without gapping: 1634
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2126
length of query: 1061
length of database: 4,897,600
effective HSP length: 100
effective length of query: 961
effective length of database: 2,051,600
effective search space: 1971587600
effective search space used: 1971587600
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC133779.6


