

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133779.2 - phase: 0
(1052 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
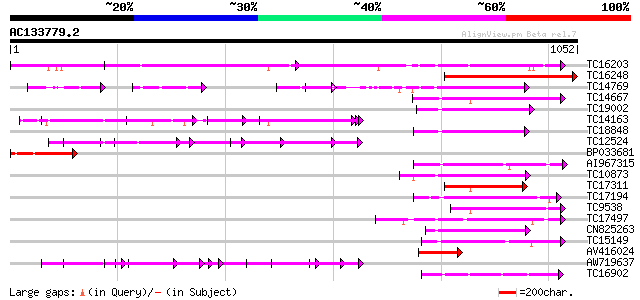
Score E
Sequences producing significant alignments: (bits) Value
TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodula... 434 e-122
TC16248 similar to UP|BAC87845 (BAC87845) Leucine-rich repeat re... 392 e-109
TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete 180 9e-46
TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, com... 164 7e-41
TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866)... 159 2e-39
TC14163 159 3e-39
TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, pa... 158 5e-39
TC12524 similar to UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kina... 157 8e-39
BP033681 156 1e-38
AI967315 155 2e-38
TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partia... 146 2e-35
TC17311 similar to UP|Q8K2Y2 (Q8K2Y2) Receptor-interacting serin... 145 2e-35
TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete 145 3e-35
TC9538 similar to UP|RLK5_ARATH (P47735) Receptor-like protein k... 142 4e-34
TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2,... 141 5e-34
CN825263 140 1e-33
TC15149 similar to UP|Q9LVI6 (Q9LVI6) Probable receptor-like pro... 140 1e-33
AV416024 137 7e-33
AW719637 132 3e-31
TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-... 129 2e-30
>TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodulation aberrant
root formation protein), complete
Length = 3308
Score = 434 bits (1117), Expect = e-122
Identities = 303/906 (33%), Positives = 476/906 (52%), Gaps = 50/906 (5%)
Frame = +2
Query: 176 IPPEIGKLNKLWFLSIQKCNLIGSIPKEIGFLTNLTLIDLSNNILSGVIPETIG-NMSKL 234
+PPEIG L KL L+I NL +P ++ LT+L ++++S+N+ SG P I M++L
Sbjct: 377 LPPEIGLLEKLENLTISMNNLTDQLPSDLASLTSLKVLNISHNLFSGQFPGNITVGMTEL 556
Query: 235 NKLYLAKNTKLYGPIPHSLWNMSSLTLIYLFNMSLSGSIPESVENLINVNELALDRNRLS 294
L N+ GP+P + + L ++L SG+IPES ++ L L+ N L+
Sbjct: 557 EALDAYDNS-FSGPLPEEIVKLEKLKYLHLAGNYFSGTIPESYSEFQSLEFLGLNANSLT 733
Query: 295 GTIPSTIGNLKNLQYLFLGM-NRLSGSIPATIGNLINLDSFSVQENNLTGTIPTTIGNLN 353
G +P ++ LK L+ L LG N G IP G++ NL + NLTG IP ++GNL
Sbjct: 734 GRVPESLAKLKTLKELHLGYSNAYEGGIPPAFGSMENLRLLEMANCNLTGEIPPSLGNLT 913
Query: 354 RLTVFEVAANKLHGRIPNGLYNITNWFSFIVSKNDFVGHLPSQICSGGLLTLLNADHNRF 413
+L V N L G IP L ++ + S +S ND G +P LTL+N N+F
Sbjct: 914 KLHSLFVQMNNLTGTIPPELSSMMSLMSLDLSINDLTGEIPESFSKLKNLTLMNFFQNKF 1093
Query: 414 TGPIPTSLKNCSSIERIRLEVNQIEGDIAQDFGVYPNLRYFDVSDNKLHGHISPNWGKSL 473
G +P+ + + ++E +++ N + + G YFDV+ N L G I P+ KS
Sbjct: 1094 RGSLPSFIGDLPNLETLQVWENNFSFVLPHNLGGNGRFLYFDVTKNHLTGLIPPDLCKSG 1273
Query: 474 NLDTF------------------------QISNNNISGVIPLELIGLTKLGRLHLSSNQF 509
L TF +++NN + G +P + L + LS+N+
Sbjct: 1274 RLKTFIITDNFFRGPIPKGIGECRSLTKIRVANNFLDGPVPPGVFQLPSVTITELSNNRL 1453
Query: 510 TGKLPKELGGMKSLFDLKLSNNHFTDSIPTEFGLLQRLEVLDLGGNELSGMIPNEVAELP 569
G+LP + G +SL L LSNN FT IP L+ L+ L L NE G IP V E+P
Sbjct: 1454 NGELPSVISG-ESLGTLTLSNNLFTGKIPAAMKNLRALQSLSLDANEFIGEIPGGVFEIP 1630
Query: 570 KLRMLNLSRNKIEGSIPSLF--RSSLASLDLSGNRLNGKIPEILGFLGQLSMLNLSHNML 627
L +N+S N + G IP+ R+SL ++DLS N L G++P+ + L LS+LNLS N +
Sbjct: 1631 MLTKVNISGNNLTGPIPTTITHRASLTAVDLSRNNLAGEVPKGMKNLMDLSILNLSRNEI 1810
Query: 628 SGTIPSFSSM--SLDFVNISNNQLEGPLPDNPAFLHAPFE-SFKNNKDLCGNFKGLDPC- 683
SG +P SL +++S+N G +P FL ++ +F N +LC + P
Sbjct: 1811 SGPVPDEIRFMTSLTTLDLSSNNFTGTVPTGGQFLVFNYDKTFAGNPNLCFPHRASCPSV 1990
Query: 684 ------GSRKSKNVLRSVLIALGALILVLFGVGISMYTLGRRKKSNEKNQTEEQTQRGVL 737
+R +R+++I + VL V ++++ + +R+ + QR
Sbjct: 1991 LYDSLRKTRAKTARVRAIVIGIALATAVLL-VAVTVHVVRKRRLHRAQAWKLTAFQRL-- 2161
Query: 738 FSIWSHDGKMMFENIIEATENFDDKYLIGVGSQGNVYKAELSSGMVVAVKKLHIITDEEI 797
++ E+++E + ++ +IG G G VY+ + +G VA+K+L +
Sbjct: 2162 --------EIKAEDVVECLK---EENIIGKGGAGIVYRGSMPNGTDVAIKRLV----GQG 2296
Query: 798 SHFSSKSFMSEIETLSGIRHRNIIKLHGFCSHSKFSFLVYKFLEGGSLGQMLNSDTQATA 857
S + F +EIETL IRHRNI++L G+ S+ + L+Y+++ GSLG+ L+ +
Sbjct: 2297 SGRNDYGFRAEIETLGKIRHRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLHG-AKGGH 2473
Query: 858 FDWEKRVNVVKGVANALSYLHHDCSPPIIHRDISSKNVLLNLDYEAQVSDFGTAKFL-KP 916
WE R + A L Y+HHDCSP IIHRD+ S N+LL+ D+EA V+DFG AKFL P
Sbjct: 2474 LRWEMRYKIAVEAARGLCYMHHDCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDP 2653
Query: 917 GL-LSWTQFAGTFGYAAPELAQTMEVNEKCDVYSFGVLALEIIVGKHP----GDLISLF- 970
G S + AG++GY APE A T++V+EK DVYSFGV+ LE+I+G+ P GD + +
Sbjct: 2654 GASQSMSSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELIIGRKPVGEFGDGVDIVG 2833
Query: 971 -----LSQSTRLMANNMLLIDVLDQRPQHVMKPVDEEVILIARLAFACLNQNPRSRPTMD 1025
+S+ ++ ++ L++ V+D P+ P+ VI + +A C+ + +RPTM
Sbjct: 2834 WVNKTMSELSQ-PSDTALVLAVVD--PRLSGYPL-TSVIHMFNIAMMCVKEMGPARPTMR 3001
Query: 1026 QVSKML 1031
+V ML
Sbjct: 3002 EVVHML 3019
Score = 247 bits (630), Expect = 8e-66
Identities = 176/591 (29%), Positives = 268/591 (44%), Gaps = 52/591 (8%)
Frame = +2
Query: 1 MIMFIILFMISWPQAVAEDSEAQALLKWKHSFDNQSQS--LLSTWKNTTNTCT--KWKGI 56
+++ L W + S+ ALLK K S L WK +T+ + G+
Sbjct: 134 LVLCFTLIWFRWTVVYSSFSDLDALLKLKESMKGAKAKHHALEDWKFSTSLSAHCSFSGV 313
Query: 57 FCDNSKSISTINLE----------NFGLKGTLHSLTFS-------------SFSNLQTLN 93
CD + + +N+ GL L +LT S S ++L+ LN
Sbjct: 314 TCDQNLRVVALNVTLVPLFGHLPPEIGLLEKLENLTISMNNLTDQLPSDLASLTSLKVLN 493
Query: 94 I-------------------------YNNYFYGTIPPQIGNISKINTLNFSLNPIDGSIP 128
I Y+N F G +P +I + K+ L+ + N G+IP
Sbjct: 494 ISHNLFSGQFPGNITVGMTELEALDAYDNSFSGPLPEEIVKLEKLKYLHLAGNYFSGTIP 673
Query: 129 QEMFTLKSLQNIDFSFCKLSGAIPNSIGNLSNLLYLDLGGNNFVGTPIPPEIGKLNKLWF 188
+ +SL+ + + L+G +P S+ L L L LG +N IPP G + L
Sbjct: 674 ESYSEFQSLEFLGLNANSLTGRVPESLAKLKTLKELHLGYSNAYEGGIPPAFGSMENLRL 853
Query: 189 LSIQKCNLIGSIPKEIGFLTNLTLIDLSNNILSGVIPETIGNMSKLNKLYLAKNTKLYGP 248
L + CNL G IP +G LT L + + N L+G IP + +M L L L+ N L G
Sbjct: 854 LEMANCNLTGEIPPSLGNLTKLHSLFVQMNNLTGTIPPELSSMMSLMSLDLSIND-LTGE 1030
Query: 249 IPHSLWNMSSLTLIYLFNMSLSGSIPESVENLINVNELALDRNRLSGTIPSTIGNLKNLQ 308
IP S + +LTL+ F GS+P + +L N+ L + N S +P +G
Sbjct: 1031IPESFSKLKNLTLMNFFQNKFRGSLPSFIGDLPNLETLQVWENNFSFVLPHNLGGNGRFL 1210
Query: 309 YLFLGMNRLSGSIPATIGNLINLDSFSVQENNLTGTIPTTIGNLNRLTVFEVAANKLHGR 368
Y + N L+G IP + L +F + +N G IP IG LT VA N L G
Sbjct: 1211YFDVTKNHLTGLIPPDLCKSGRLKTFIITDNFFRGPIPKGIGECRSLTKIRVANNFLDGP 1390
Query: 369 IPNGLYNITNWFSFIVSKNDFVGHLPSQICSGGLLTLLNADHNRFTGPIPTSLKNCSSIE 428
+P G++ + + +S N G LPS I SG L L +N FTG IP ++KN +++
Sbjct: 1391VPPGVFQLPSVTITELSNNRLNGELPSVI-SGESLGTLTLSNNLFTGKIPAAMKNLRALQ 1567
Query: 429 RIRLEVNQIEGDIAQDFGVYPNLRYFDVSDNKLHGHISPNWGKSLNLDTFQISNNNISGV 488
+ L+ N+ G+I P L ++S N L G I +L +S NN++G
Sbjct: 1568SLSLDANEFIGEIPGGVFEIPMLTKVNISGNNLTGPIPTTITHRASLTAVDLSRNNLAGE 1747
Query: 489 IPLELIGLTKLGRLHLSSNQFTGKLPKELGGMKSLFDLKLSNNHFTDSIPT 539
+P + L L L+LS N+ +G +P E+ M SL L LS+N+FT ++PT
Sbjct: 1748VPKGMKNLMDLSILNLSRNEISGPVPDEIRFMTSLTTLDLSSNNFTGTVPT 1900
>TC16248 similar to UP|BAC87845 (BAC87845) Leucine-rich repeat receptor-like
protein kinase 1, partial (17%)
Length = 866
Score = 392 bits (1006), Expect = e-109
Identities = 194/247 (78%), Positives = 215/247 (86%), Gaps = 1/247 (0%)
Frame = +2
Query: 807 SEIETLSGIRHRNIIKLHGFCSHSKFSFLVYKFLEGGSLGQMLNSDTQATAFDWEKRVNV 866
SEI+TL+ IRHRN+IKLHGFC HSKFSFLVY+FLEGGSL Q+LNSDTQA AF WEKRVNV
Sbjct: 2 SEIQTLTEIRHRNVIKLHGFCLHSKFSFLVYEFLEGGSLDQVLNSDTQAAAFHWEKRVNV 181
Query: 867 VKGVANALSYLHHDCSPPIIHRDISSKNVLLNLDYEAQVSDFGTAKFLKPGLLSWTQFAG 926
VKGVANALSY+HHDCSPPIIHRDISSKNVLL+L YEA VSDFGTAKFLKPG +WT FAG
Sbjct: 182 VKGVANALSYMHHDCSPPIIHRDISSKNVLLDLQYEAHVSDFGTAKFLKPGSHTWTAFAG 361
Query: 927 TFGYAAPELAQTMEVNEKCDVYSFGVLALEIIVGKHPGDLISLFLSQSTRLMANNMLLID 986
TFGYAAPELAQTM+VNEKCDVYSFGV ALEII+G HPGDLIS +S ST M N++LLID
Sbjct: 362 TFGYAAPELAQTMQVNEKCDVYSFGVFALEIIMGMHPGDLISSLMSPSTVPMVNDLLLID 541
Query: 987 VLDQRPQHVMKPVDEEVILIARLAFACLNQNPRSRPTMDQVSKMLAIGKSPL-VGMQLHM 1045
+LDQRP V KP+DEEVILIA LA ACL +NP SRPTMDQVSK L +GK PL +G Q M
Sbjct: 542 ILDQRPHQVEKPIDEEVILIASLALACLRKNPHSRPTMDQVSKALVLGKPPLALGYQFPM 721
Query: 1046 IRLGQLN 1052
+R+GQL+
Sbjct: 722 VRVGQLH 742
>TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete
Length = 3495
Score = 180 bits (457), Expect = 9e-46
Identities = 144/434 (33%), Positives = 210/434 (48%), Gaps = 20/434 (4%)
Frame = +3
Query: 550 LDLGGNELSGMIPNEVAELPKLRMLNLSRNKIEGSIPSLFRSS-LASLDLSGNRLNGKIP 608
LDL + L G+IP+ +AE+ L LN+S N +GS+PS SS L S+DLS N L GK+P
Sbjct: 1608 LDLSSSNLKGLIPSSIAEMTNLETLNISHNSFDGSVPSFPLSSLLISVDLSYNDLMGKLP 1787
Query: 609 EILGFLGQLSMLNLSHNMLSGTIPSFSSMSLDFVNISNNQLEGPLPDNPAFLHAPFESFK 668
E + L L L N E P++PA ++ S
Sbjct: 1788 ESIVKLPHLKSLYFGCN------------------------EHMSPEDPANMN----SSL 1883
Query: 669 NNKDLCGNFKGLDPCGSRKSKNVLRSVLIALGALILVLFGVGISMYTLGRRKK------- 721
N D G KG + SR + ++ + LI + FGV ++ R+K
Sbjct: 1884 INTDY-GRCKGKE---SRFGQVIVIGAITCGSLLITLAFGV---LFVCRYRQKLIPWEGF 2042
Query: 722 SNEKNQTEEQTQRGVLFSIWSHDG---------KMMFENIIEATENFDDKYLIGVGSQGN 772
+ +K E ++FS+ S D E I ATE + K LIG G G+
Sbjct: 2043 AGKKYPME----TNIIFSLPSKDDFFIKSVSIQAFTLEYIEVATERY--KTLIGEGGFGS 2204
Query: 773 VYKAELSSGMVVAVKKLHIITDEEISHFSSKSFMSEIETLSGIRHRNIIKLHGFCSHSKF 832
VY+ L+ G VAVK S ++ F +E+ LS I+H N++ L G+C+ S
Sbjct: 2205 VYRGTLNDGQEVAVK-----VRSSTSTQGTREFDNELNLLSAIQHENLVPLLGYCNESDQ 2369
Query: 833 SFLVYKFLEGGSLGQMLNSD-TQATAFDWEKRVNVVKGVANALSYLHHDCSPPIIHRDIS 891
LVY F+ GSL L + + DW R+++ G A L+YLH +IHRDI
Sbjct: 2370 QILVYPFMSNGSLQDRLYGEPAKRKILDWPTRLSIALGAARGLAYLHTFPGRSVIHRDIK 2549
Query: 892 SKNVLLNLDYEAQVSDFGTAKFLKPGLLSWT--QFAGTFGYAAPELAQTMEVNEKCDVYS 949
S N+LL+ A+V+DFG +K+ S+ + GT GY PE +T +++EK DV+S
Sbjct: 2550 SSNILLDHSMCAKVADFGFSKYAPQEGDSYVSLEVRGTAGYLDPEYYKTQQLSEKSDVFS 2729
Query: 950 FGVLALEIIVGKHP 963
FGV+ LEI+ G+ P
Sbjct: 2730 FGVVLLEIVSGREP 2771
Score = 59.3 bits (142), Expect = 3e-09
Identities = 48/145 (33%), Positives = 60/145 (41%), Gaps = 1/145 (0%)
Frame = +3
Query: 34 NQSQSLLSTWKNTTNTCTKWKGIFCDNSKSISTINLENFGLKGTLHSLTFSSFSNLQTLN 93
N L +W WKGI CD S S I L SS SNL+
Sbjct: 1500 NSGNRALESWSGDPCILLPWKGIACDGSNGSSVIT-----------KLDLSS-SNLK--- 1634
Query: 94 IYNNYFYGTIPPQIGNISKINTLNFSLNPIDGSIPQEMFTLKSLQ-NIDFSFCKLSGAIP 152
G IP I ++ + TLN S N DGS+P F L SL ++D S+ L G +P
Sbjct: 1635 -------GLIPSSIAEMTNLETLNISHNSFDGSVPS--FPLSSLLISVDLSYNDLMGKLP 1787
Query: 153 NSIGNLSNLLYLDLGGNNFVGTPIP 177
SI L +L L G N + P
Sbjct: 1788 ESIVKLPHLKSLYFGCNEHMSPEDP 1862
Score = 54.3 bits (129), Expect = 1e-07
Identities = 37/113 (32%), Positives = 58/113 (50%), Gaps = 1/113 (0%)
Frame = +3
Query: 495 GLTKLGRLHLSSNQFTGKLPKELGGMKSLFDLKLSNNHFTDSIPTEFGLLQRLEVLDLGG 554
G + + +L LSS+ G +P + M +L L +S+N F S+P+ F L L +DL
Sbjct: 1587 GSSVITKLDLSSSNLKGLIPSSIAEMTNLETLNISHNSFDGSVPS-FPLSSLLISVDLSY 1763
Query: 555 NELSGMIPNEVAELPKLRMLNLSRNK-IEGSIPSLFRSSLASLDLSGNRLNGK 606
N+L G +P + +LP L+ L N+ + P+ SSL + D R GK
Sbjct: 1764 NDLMGKLPESIVKLPHLKSLYFGCNEHMSPEDPANMNSSLINTDY--GRCKGK 1916
Score = 42.4 bits (98), Expect = 4e-04
Identities = 37/138 (26%), Positives = 63/138 (44%), Gaps = 1/138 (0%)
Frame = +3
Query: 228 IGNMSKLNKLYLAKNTKLYGPIPHSLWNMSSLTLIYLFNMSLSGSIPESVENLINVNELA 287
+G + K+ + L +N+ G W+ L+ ++ GS SV + +L
Sbjct: 1458 VGVIQKMREELLLQNS---GNRALESWSGDPCILLPWKGIACDGSNGSSV-----ITKLD 1613
Query: 288 LDRNRLSGTIPSTIGNLKNLQYLFLGMNRLSGSIPA-TIGNLINLDSFSVQENNLTGTIP 346
L + L G IPS+I + NL+ L + N GS+P+ + +L L S + N+L G +P
Sbjct: 1614 LSSSNLKGLIPSSIAEMTNLETLNISHNSFDGSVPSFPLSSL--LISVDLSYNDLMGKLP 1787
Query: 347 TTIGNLNRLTVFEVAANK 364
+I L L N+
Sbjct: 1788 ESIVKLPHLKSLYFGCNE 1841
Score = 40.8 bits (94), Expect = 0.001
Identities = 32/95 (33%), Positives = 45/95 (46%)
Frame = +3
Query: 164 LDLGGNNFVGTPIPPEIGKLNKLWFLSIQKCNLIGSIPKEIGFLTNLTLIDLSNNILSGV 223
LDL +N G IP I ++ L L+I + GS+P + L +DLS N L G
Sbjct: 1608 LDLSSSNLKGL-IPSSIAEMTNLETLNISHNSFDGSVPS-FPLSSLLISVDLSYNDLMGK 1781
Query: 224 IPETIGNMSKLNKLYLAKNTKLYGPIPHSLWNMSS 258
+PE+I + L LY N + P NM+S
Sbjct: 1782 LPESIVKLPHLKSLYFGCNEHM---SPEDPANMNS 1877
Score = 35.8 bits (81), Expect = 0.036
Identities = 26/93 (27%), Positives = 43/93 (45%), Gaps = 2/93 (2%)
Frame = +3
Query: 384 VSKNDFVGHLPSQICSGGLLTLLNADHNRFTGPIPTSLKNCSSIERIRLEVNQIEGDIAQ 443
+S ++ G +PS I L LN HN F G +P S S + + L N + G + +
Sbjct: 1614 LSSSNLKGLIPSSIAEMTNLETLNISHNSFDGSVP-SFPLSSLLISVDLSYNDLMGKLPE 1790
Query: 444 DFGVYPNLR--YFDVSDNKLHGHISPNWGKSLN 474
P+L+ YF ++ H+SP ++N
Sbjct: 1791 SIVKLPHLKSLYFGCNE-----HMSPEDPANMN 1874
>TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, complete
Length = 1591
Score = 164 bits (415), Expect = 7e-41
Identities = 104/295 (35%), Positives = 151/295 (50%), Gaps = 10/295 (3%)
Frame = +2
Query: 747 MMFENIIEATENFDDKYLIGVGSQGNVYKAELSSGMVVAVKKLHIITDEEISHFSSKSFM 806
+ + + E T+NF K LIG GS G VY A L+ G VAVKKL + ++ E ++ F+
Sbjct: 323 LSLDELKEKTDNFGSKALIGEGSYGRVYYATLNDGNAVAVKKLDVSSEPE----TNNEFL 490
Query: 807 SEIETLSGIRHRNIIKLHGFCSHSKFSFLVYKFLEGGSLGQMLNSDT------QATAFDW 860
+++ +S +++ N ++LHG+C L Y+F GSL +L+ DW
Sbjct: 491 TQVSMVSRLKNDNFVELHGYCVEGNLRVLAYEFATMGSLHDILHGRKGVQGAQPGPTLDW 670
Query: 861 EKRVNVVKGVANALSYLHHDCSPPIIHRDISSKNVLLNLDYEAQVSDFGTAKFL--KPGL 918
+RV + A L YLH P IIHRDI S NVL+ DY+A+++DF +
Sbjct: 671 IQRVRIAVDAARGLEYLHEKVQPAIIHRDIRSSNVLIFEDYKAKIADFNLSNQAPDMAAR 850
Query: 919 LSWTQFAGTFGYAAPELAQTMEVNEKCDVYSFGVLALEIIVGKHPGDLISLFLSQSTRLM 978
L T+ GTFGY APE A T ++ +K DVYSFGV+ LE++ G+ P D QS
Sbjct: 851 LHSTRVLGTFGYHAPEYAMTGQLTQKSDVYSFGVVLLELLTGRKPVDHTMPRGQQSLVTW 1030
Query: 979 ANNMLLIDVLDQ--RPQHVMKPVDEEVILIARLAFACLNQNPRSRPTMDQVSKML 1031
A L D + Q P+ + + V +A +A C+ RP M V K L
Sbjct: 1031 ATPRLSEDKVKQCVDPKLKGEYPPKGVAKLAAVAALCVQYEAEFRPNMSIVVKAL 1195
>TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866), partial
(48%)
Length = 780
Score = 159 bits (403), Expect = 2e-39
Identities = 85/222 (38%), Positives = 131/222 (58%), Gaps = 2/222 (0%)
Frame = +1
Query: 755 ATENFDDKYLIGVGSQGNVYKAELSSGMVVAVKKLHIITDEEISHFSSKSFMSEIETLSG 814
AT NF+ +G G G+VY +L G +AVK+L + +++ + F E+E L+
Sbjct: 52 ATNNFNYDNKLGEGGFGSVYWGQLWDGSQIAVKRLKVWSNK-----ADMEFAVEVEILAR 216
Query: 815 IRHRNIIKLHGFCSHSKFSFLVYKFLEGGSLGQMLNSDTQATAF-DWEKRVNVVKGVANA 873
+RH+N++ L G+C+ + +VY ++ SL L+ + DW +R+N+ G A
Sbjct: 217 VRHKNLLSLRGYCAEGQERLIVYDYMPNLSLLSHLHGQHSSECLLDWNRRMNIAIGSAEG 396
Query: 874 LSYLHHDCSPPIIHRDISSKNVLLNLDYEAQVSDFGTAKFLKPGLLS-WTQFAGTFGYAA 932
+ YLHH +P IIHRDI + NVLL+ D++A+V+DFG AK + G T+ GT GY A
Sbjct: 397 IVYLHHQATPHIIHRDIKASNVLLDSDFQARVADFGFAKLIPDGATHVTTRVKGTLGYLA 576
Query: 933 PELAQTMEVNEKCDVYSFGVLALEIIVGKHPGDLISLFLSQS 974
PE A + NE CDV+SFG+L LE+ GK P + +S + +S
Sbjct: 577 PEYAMLGKANECCDVFSFGILLLELASGKKPLEKLSSTVKRS 702
>TC14163
Length = 1712
Score = 159 bits (401), Expect = 3e-39
Identities = 128/427 (29%), Positives = 187/427 (42%), Gaps = 2/427 (0%)
Frame = +1
Query: 218 NILSGVIPETIGNMSKLNKLYLAKNTKLYGPIPHSLWNMSSLTLIYLFNMSLSGSIPESV 277
+ SG I ++ + L+ YL + GP P L+ + L IY+ N LSG IPE++
Sbjct: 370 SFFSGTISPSLSKIKNLDGFYLLNLKNISGPFPGFLFKLPKLQFIYIENNQLSGRIPENI 549
Query: 278 ENLINVNELALDRNRLSGTIPSTIGNLKNLQYLFLGMNRLSGSIPATIGNLINLDSFSVQ 337
NL ++ L+L NR +GTIPS++G L +L L LG N L+G+IPATI L NL S++
Sbjct: 550 GNLTRLDVLSLTGNRFTGTIPSSVGGLTHLTQLQLGNNSLTGTIPATIARLKNLTYLSLE 729
Query: 338 ENNLTGTIPTTIGNLNRLTVFEVAANKLHGRIPNGLYNITNWFSFI-VSKNDFVGHLPSQ 396
N +G IP + L + ++ NK G+IP + + ++ + N G +P
Sbjct: 730 GNQFSGAIPDFFSSFTDLGILRLSRNKFSGKIPASISTLAPKLRYLELGHNQLSGKIPDF 909
Query: 397 ICSGGLLTLLNADHNRFTGPIPTSLKNCSSIERIRLEVNQIEGDIAQDFGVYPNLRYFDV 456
+ L L+ NRF+G +P S KN + I + L N + D + V + D+
Sbjct: 910 LGKFRALDTLDLSSNRFSGTVPASFKNLTKIFNLNL-ANNLLVDPFPEMNV-KGIESLDL 1083
Query: 457 SDNKLHGHISPNWGKSLNLDTFQISNNNISGVIPLELIGLTKLGRLHLSSNQFTGKLPKE 516
S+N H + P W V +I KL R
Sbjct: 1084SNNMFHLNQIPKW------------------VTSSPIIFSLKLAR--------------- 1164
Query: 517 LGGMKSLFDLKLSNNHFTDSIPTEFGLLQRLEVLDLGGNELSGMIPNEVAELPKLRMLNL 576
G L D K + +F D I DL GNE+SG V L
Sbjct: 1165CGIKMKLDDWKPAETYFYDFI-------------DLSGNEISGSAVGLVNSTEYLVGFWG 1305
Query: 577 SRNKIEGSIPSL-FRSSLASLDLSGNRLNGKIPEILGFLGQLSMLNLSHNMLSGTIPSFS 635
S NK++ L F LDLS N + GK+P+ + L LN+S+N L G IP
Sbjct: 1306SGNKLKFDFERLKFGERFKYLDLSHNLVFGKVPK---SVAGLEKLNVSYNHLCGEIPKTK 1476
Query: 636 SMSLDFV 642
+ FV
Sbjct: 1477FPASAFV 1497
Score = 132 bits (332), Expect = 3e-31
Identities = 86/223 (38%), Positives = 115/223 (51%), Gaps = 30/223 (13%)
Frame = +1
Query: 463 GHISPNWGKSLNLDTFQ-------------------------ISNNNISGVIPLELIGLT 497
G ISP+ K NLD F I NN +SG IP + LT
Sbjct: 382 GTISPSLSKIKNLDGFYLLNLKNISGPFPGFLFKLPKLQFIYIENNQLSGRIPENIGNLT 561
Query: 498 KLGRLHLSSNQFTGKLPKELGGMKSLFDLKLSNNHFTDSIPTEFGLLQRLEVLDLGGNEL 557
+L L L+ N+FTG +P +GG+ L L+L NN T +IP L+ L L L GN+
Sbjct: 562 RLDVLSLTGNRFTGTIPSSVGGLTHLTQLQLGNNSLTGTIPATIARLKNLTYLSLEGNQF 741
Query: 558 SGMIPNEVAELPKLRMLNLSRNKIEGSIP---SLFRSSLASLDLSGNRLNGKIPEILGFL 614
SG IP+ + L +L LSRNK G IP S L L+L N+L+GKIP+ LG
Sbjct: 742 SGAIPDFFSSFTDLGILRLSRNKFSGKIPASISTLAPKLRYLELGHNQLSGKIPDFLGKF 921
Query: 615 GQLSMLNLSHNMLSGTIP-SFSSMSLDF-VNISNNQLEGPLPD 655
L L+LS N SGT+P SF +++ F +N++NN L P P+
Sbjct: 922 RALDTLDLSSNRFSGTVPASFKNLTKIFNLNLANNLLVDPFPE 1050
Score = 121 bits (304), Expect = 5e-28
Identities = 122/448 (27%), Positives = 200/448 (44%), Gaps = 25/448 (5%)
Frame = +1
Query: 19 DSEAQALLKWKHSFDNQSQSLLSTWKNTTNTCTKWKGIFC-DNSKSISTI-------NLE 70
D EA L+ +K + +L +W T+ CT W+G+ C + K ++++ N +
Sbjct: 196 DDEA-GLMGFKSGIKSDPSGILKSWIPGTDCCT-WQGVTCLFDDKRVTSLYLSGNPENPK 369
Query: 71 NFGLKGTLHSLTFSSFSNLQTLNIYN-NYFYGTIPPQIGNISKINTLNFSLNPIDGSIPQ 129
+F GT+ S + S NL + N G P + + K+ + N + G IP+
Sbjct: 370 SF-FSGTI-SPSLSKIKNLDGFYLLNLKNISGPFPGFLFKLPKLQFIYIENNQLSGRIPE 543
Query: 130 EMFTLKSLQNIDFSFCKLSGAIPNSIGNLSNLLYLDLGGNNFVGTPIPPEIGKLNKLWFL 189
+ L L + + + +G IP+S+G L++L L LG N+ GT IP I +L L +L
Sbjct: 544 NIGNLTRLDVLSLTGNRFTGTIPSSVGGLTHLTQLQLGNNSLTGT-IPATIARLKNLTYL 720
Query: 190 SIQKCNLIGSIPKEIGFLTNLTLIDLSNNILSGVIPETIGNMS-KLNKLYLAKNTKLYGP 248
S++ G+IP T+L ++ LS N SG IP +I ++ KL L L N +L G
Sbjct: 721 SLEGNQFSGAIPDFFSSFTDLGILRLSRNKFSGKIPASISTLAPKLRYLELGHN-QLSGK 897
Query: 249 IPHSLWNMSSLTLIYLFNMSLSGSIPESVENLINVNELALDRNRLSGTIPSTIGNLKNLQ 308
IP L +L + L + SG++P S +NL + L L N L P N+K ++
Sbjct: 898 IPDFLGKFRALDTLDLSSNRFSGTVPASFKNLTKIFNLNLANNLLVDPFPEM--NVKGIE 1071
Query: 309 YLFLGMNRLS-GSIP--------------ATIGNLINLDSFSVQENNLTGTIPTTIGNLN 353
L L N IP A G + LD + E I + GN
Sbjct: 1072SLDLSNNMFHLNQIPKWVTSSPIIFSLKLARCGIKMKLDDWKPAETYFYDFIDLS-GN-- 1242
Query: 354 RLTVFEVAANKLHGRIPNGLYNITNWFSFIVSKNDFVGHLPSQICSGGLLTLLNADHNRF 413
E++ + + G + + Y + W S K DF ++ G L+ HN
Sbjct: 1243-----EISGSAV-GLVNSTEYLVGFWGSGNKLKFDF-----ERLKFGERFKYLDLSHNLV 1389
Query: 414 TGPIPTSLKNCSSIERIRLEVNQIEGDI 441
G +P K+ + +E++ + N + G+I
Sbjct: 1390FGKVP---KSVAGLEKLNVSYNHLCGEI 1464
Score = 109 bits (272), Expect = 3e-24
Identities = 79/291 (27%), Positives = 132/291 (45%), Gaps = 6/291 (2%)
Frame = +1
Query: 367 GRIPNGLYNITNWFSF-IVSKNDFVGHLPSQICSGGLLTLLNADHNRFTGPIPTSLKNCS 425
G I L I N F +++ + G P + L + ++N+ +G IP ++ N +
Sbjct: 382 GTISPSLSKIKNLDGFYLLNLKNISGPFPGFLFKLPKLQFIYIENNQLSGRIPENIGNLT 561
Query: 426 SIERIRLEVNQIEGDIAQDFGVYPNLRYFDVSDNKLHGHISPNWGKSLNLDTFQISNNNI 485
++ + L N+ G I G +L + +N L G I + NL + N
Sbjct: 562 RLDVLSLTGNRFTGTIPSSVGGLTHLTQLQLGNNSLTGTIPATIARLKNLTYLSLEGNQF 741
Query: 486 SGVIPLELIGLTKLGRLHLSSNQFTGKLPKELGGMK-SLFDLKLSNNHFTDSIPTEFGLL 544
SG IP T LG L LS N+F+GK+P + + L L+L +N + IP G
Sbjct: 742 SGAIPDFFSSFTDLGILRLSRNKFSGKIPASISTLAPKLRYLELGHNQLSGKIPDFLGKF 921
Query: 545 QRLEVLDLGGNELSGMIPNEVAELPKLRMLNLSRNKIEGSIPSLFRSSLASLDLSGNRLN 604
+ L+ LDL N SG +P L K+ LNL+ N + P + + SLDLS N +
Sbjct: 922 RALDTLDLSSNRFSGTVPASFKNLTKIFNLNLANNLLVDPFPEMNVKGIESLDLSNNMFH 1101
Query: 605 -GKIPEILGFLGQLSMLNLSHNMLSGTIPSF---SSMSLDFVNISNNQLEG 651
+IP+ + + L L+ + + + + DF+++S N++ G
Sbjct: 1102LNQIPKWVTSSPIIFSLKLARCGIKMKLDDWKPAETYFYDFIDLSGNEISG 1254
Score = 84.3 bits (207), Expect = 9e-17
Identities = 86/316 (27%), Positives = 130/316 (40%), Gaps = 27/316 (8%)
Frame = +1
Query: 60 NSKSISTINLENFGLKGTLHSLTFSSFSNLQTLNIYNNYFYGTIPPQIGNISKINTLNFS 119
N + ++L GT+ S + ++L L + NN GTIP I + + L+
Sbjct: 553 NLTRLDVLSLTGNRFTGTIPS-SVGGLTHLTQLQLGNNSLTGTIPATIARLKNLTYLSLE 729
Query: 120 LNPIDGSIPQEMFTLKSLQNIDFSFCKLSGAIPNSIGNLS-NLLYLDLGGNNFVGTPIPP 178
N G+IP + L + S K SG IP SI L+ L YL+LG N G IP
Sbjct: 730 GNQFSGAIPDFFSSFTDLGILRLSRNKFSGKIPASISTLAPKLRYLELGHNQLSG-KIPD 906
Query: 179 EIGKLNKLWFLSIQKCNLIGSIPKEIGFLTNLTLIDLSNNILSGVIPETIGNMSKLNKLY 238
+GK L L + G++P LT + ++L+NN+L PE N+ + L
Sbjct: 907 FLGKFRALDTLDLSSNRFSGTVPASFKNLTKIFNLNLANNLLVDPFPEM--NVKGIESLD 1080
Query: 239 LAKNTKLYGPIPHSLWNMSSLTLI---------------------YLFN-MSLSGS-IPE 275
L+ N IP W SS + Y ++ + LSG+ I
Sbjct: 1081LSNNMFHLNQIPK--WVTSSPIIFSLKLARCGIKMKLDDWKPAETYFYDFIDLSGNEISG 1254
Query: 276 SVENLINVNELALDRNRLSGTIPSTIGNLK---NLQYLFLGMNRLSGSIPATIGNLINLD 332
S L+N E + + LK +YL L N + G +P ++ L+
Sbjct: 1255SAVGLVNSTEYLVGFWGSGNKLKFDFERLKFGERFKYLDLSHNLVFGKVPKSVA---GLE 1425
Query: 333 SFSVQENNLTGTIPTT 348
+V N+L G IP T
Sbjct: 1426KLNVSYNHLCGEIPKT 1473
>TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, partial (64%)
Length = 969
Score = 158 bits (399), Expect = 5e-39
Identities = 84/217 (38%), Positives = 123/217 (55%), Gaps = 2/217 (0%)
Frame = +2
Query: 749 FENIIEATENFDDKYLIGVGSQGNVYKAELSSGMVVAVKKLHIITDEEISHFSSKSFMSE 808
++ + AT F D +G G G+VY S G+ +AVKKL + + + F E
Sbjct: 272 YKELHAATGGFSDDNKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSK-----AEMEFAVE 436
Query: 809 IETLSGIRHRNIIKLHGFCSHSKFSFLVYKFLEGGSLGQMLNSDTQATA-FDWEKRVNVV 867
+E L +RH+N++ L G+C +VY ++ SL L+ +W+KR+ +
Sbjct: 437 VEVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQFAVEVQLNWQKRMKIA 616
Query: 868 KGVANALSYLHHDCSPPIIHRDISSKNVLLNLDYEAQVSDFGTAKFLKPGLLSW-TQFAG 926
G A + YLHH+ +P IIHRDI + NVLLN D+E V+DFG AK + G+ T+ G
Sbjct: 617 IGSAEGILYLHHEVTPHIIHRDIKASNVLLNSDFEPLVADFGFAKLIPEGVSHMTTRVKG 796
Query: 927 TFGYAAPELAQTMEVNEKCDVYSFGVLALEIIVGKHP 963
T GY APE A +V+E CDVYSFG+L LE++ G+ P
Sbjct: 797 TLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKP 907
>TC12524 similar to UP|Q9LKZ6 (Q9LKZ6) Receptor-like protein kinase 1,
partial (27%)
Length = 834
Score = 157 bits (397), Expect = 8e-39
Identities = 104/318 (32%), Positives = 148/318 (45%)
Frame = +2
Query: 195 NLIGSIPKEIGFLTNLTLIDLSNNILSGVIPETIGNMSKLNKLYLAKNTKLYGPIPHSLW 254
N G IP EIG LT L D + LSG IP +G + KL+ L+L N
Sbjct: 26 NYQGGIPPEIGNLTQLLRFDAAYCGLSGEIPAELGKLQKLDTLFLQVNV----------- 172
Query: 255 NMSSLTLIYLFNMSLSGSIPESVENLINVNELALDRNRLSGTIPSTIGNLKNLQYLFLGM 314
LSGS+ + +L ++ + L N LSG +P++ LKNL L L
Sbjct: 173 --------------LSGSLTPELGHLKSLKSMDLSNNMLSGQVPASFAELKNLTLLNLFR 310
Query: 315 NRLSGSIPATIGNLINLDSFSVQENNLTGTIPTTIGNLNRLTVFEVAANKLHGRIPNGLY 374
NRL G+IP +G + L+ + ENN TG+IP ++G +LT+ ++++NKL
Sbjct: 311 NRLHGAIPEFVGEMPALEVLQLWENNFTGSIPQSLGKNGKLTLVDLSSNKL--------- 463
Query: 375 NITNWFSFIVSKNDFVGHLPSQICSGGLLTLLNADHNRFTGPIPTSLKNCSSIERIRLEV 434
G LP +CSG L L A N GPIP SL C S+ RIR+
Sbjct: 464 ---------------TGTLPPHMCSGNRLQTLIALGNFLFGPIPESLGKCESLTRIRMGQ 598
Query: 435 NQIEGDIAQDFGVYPNLRYFDVSDNKLHGHISPNWGKSLNLDTFQISNNNISGVIPLELI 494
N + G I + P L + DN L G S N+ +SNN +SG +P +
Sbjct: 599 NFLNGSIPKGLFGLPKLTQVEFQDNLLSGEFPETGSVSHNIGQITLSNNKLSGPLPSTIG 778
Query: 495 GLTKLGRLHLSSNQFTGK 512
T + +L L N+F+G+
Sbjct: 779 NFTSMQKLLLDGNKFSGR 832
Score = 148 bits (373), Expect = 5e-36
Identities = 89/243 (36%), Positives = 136/243 (55%)
Frame = +2
Query: 101 GTIPPQIGNISKINTLNFSLNPIDGSIPQEMFTLKSLQNIDFSFCKLSGAIPNSIGNLSN 160
G IP ++G + K++TL +N + GS+ E+ LKSL+++D S LSG +P S L N
Sbjct: 107 GEIPAELGKLQKLDTLFLQVNVLSGSLTPELGHLKSLKSMDLSNNMLSGQVPASFAELKN 286
Query: 161 LLYLDLGGNNFVGTPIPPEIGKLNKLWFLSIQKCNLIGSIPKEIGFLTNLTLIDLSNNIL 220
L L+L N G IP +G++ L L + + N GSIP+ +G LTL+DLS+N L
Sbjct: 287 LTLLNLFRNRLHGA-IPEFVGEMPALEVLQLWENNFTGSIPQSLGKNGKLTLVDLSSNKL 463
Query: 221 SGVIPETIGNMSKLNKLYLAKNTKLYGPIPHSLWNMSSLTLIYLFNMSLSGSIPESVENL 280
+G +P + + ++L L +A L+GPIP SL SLT I + L+GSIP+ + L
Sbjct: 464 TGTLPPHMCSGNRLQTL-IALGNFLFGPIPESLGKCESLTRIRMGQNFLNGSIPKGLFGL 640
Query: 281 INVNELALDRNRLSGTIPSTIGNLKNLQYLFLGMNRLSGSIPATIGNLINLDSFSVQENN 340
+ ++ N LSG P T N+ + L N+LSG +P+TIGN ++ + N
Sbjct: 641 PKLTQVEFQDNLLSGEFPETGSVSHNIGQITLSNNKLSGPLPSTIGNFTSMQKLLLDGNK 820
Query: 341 LTG 343
+G
Sbjct: 821 FSG 829
Score = 139 bits (349), Expect = 3e-33
Identities = 85/271 (31%), Positives = 137/271 (50%)
Frame = +2
Query: 169 NNFVGTPIPPEIGKLNKLWFLSIQKCNLIGSIPKEIGFLTNLTLIDLSNNILSGVIPETI 228
NN+ G IPPEIG L +L C L G IP E+G L L + L N+LSG + +
Sbjct: 23 NNYQGG-IPPEIGNLTQLLRFDAAYCGLSGEIPAELGKLQKLDTLFLQVNVLSGSLTPEL 199
Query: 229 GNMSKLNKLYLAKNTKLYGPIPHSLWNMSSLTLIYLFNMSLSGSIPESVENLINVNELAL 288
G++ L + L+ N L G +P S + +LTL+ LF L G+IPE V + + L L
Sbjct: 200 GHLKSLKSMDLS-NNMLSGQVPASFAELKNLTLLNLFRNRLHGAIPEFVGEMPALEVLQL 376
Query: 289 DRNRLSGTIPSTIGNLKNLQYLFLGMNRLSGSIPATIGNLINLDSFSVQENNLTGTIPTT 348
N +G+IP ++G L + L N+L+G++P + + L + N L G IP +
Sbjct: 377 WENNFTGSIPQSLGKNGKLTLVDLSSNKLTGTLPPHMCSGNRLQTLIALGNFLFGPIPES 556
Query: 349 IGNLNRLTVFEVAANKLHGRIPNGLYNITNWFSFIVSKNDFVGHLPSQICSGGLLTLLNA 408
+G LT + N L+G IP GL+ + N G P + +
Sbjct: 557 LGKCESLTRIRMGQNFLNGSIPKGLFGLPKLTQVEFQDNLLSGEFPETGSVSHNIGQITL 736
Query: 409 DHNRFTGPIPTSLKNCSSIERIRLEVNQIEG 439
+N+ +GP+P+++ N +S++++ L+ N+ G
Sbjct: 737 SNNKLSGPLPSTIGNFTSMQKLLLDGNKFSG 829
Score = 132 bits (333), Expect = 2e-31
Identities = 79/247 (31%), Positives = 127/247 (50%)
Frame = +2
Query: 73 GLKGTLHSLTFSSFSNLQTLNIYNNYFYGTIPPQIGNISKINTLNFSLNPIDGSIPQEMF 132
GL G + + L TL + N G++ P++G++ + +++ S N + G +P
Sbjct: 98 GLSGEIPA-ELGKLQKLDTLFLQVNVLSGSLTPELGHLKSLKSMDLSNNMLSGQVPASFA 274
Query: 133 TLKSLQNIDFSFCKLSGAIPNSIGNLSNLLYLDLGGNNFVGTPIPPEIGKLNKLWFLSIQ 192
LK+L ++ +L GAIP +G + L L L NNF G+ IP +GK KL + +
Sbjct: 275 ELKNLTLLNLFRNRLHGAIPEFVGEMPALEVLQLWENNFTGS-IPQSLGKNGKLTLVDLS 451
Query: 193 KCNLIGSIPKEIGFLTNLTLIDLSNNILSGVIPETIGNMSKLNKLYLAKNTKLYGPIPHS 252
L G++P + L + N L G IPE++G L ++ + +N L G IP
Sbjct: 452 SNKLTGTLPPHMCSGNRLQTLIALGNFLFGPIPESLGKCESLTRIRMGQNF-LNGSIPKG 628
Query: 253 LWNMSSLTLIYLFNMSLSGSIPESVENLINVNELALDRNRLSGTIPSTIGNLKNLQYLFL 312
L+ + LT + + LSG PE+ N+ ++ L N+LSG +PSTIGN ++Q L L
Sbjct: 629 LFGLPKLTQVEFQDNLLSGEFPETGSVSHNIGQITLSNNKLSGPLPSTIGNFTSMQKLLL 808
Query: 313 GMNRLSG 319
N+ SG
Sbjct: 809 DGNKFSG 829
Score = 124 bits (312), Expect = 6e-29
Identities = 80/249 (32%), Positives = 126/249 (50%), Gaps = 4/249 (1%)
Frame = +2
Query: 410 HNRFTGPIPTSLKNCSSIERIRLEVNQIEGDIAQDFGVYPNLRYFDVSDNKLHGHISPNW 469
+N + G IP + N + + R + G+I + G L + N L G ++P
Sbjct: 20 YNNYQGGIPPEIGNLTQLLRFDAAYCGLSGEIPAELGKLQKLDTLFLQVNVLSGSLTPEL 199
Query: 470 GKSLNLDTFQISNNNISGVIPLELIGLTKLGRLHLSSNQFTGKLPKELGGMKSLFDLKLS 529
G +L + +SNN +SG +P L L L+L N+ G +P+ +G M +L L+L
Sbjct: 200 GHLKSLKSMDLSNNMLSGQVPASFAELKNLTLLNLFRNRLHGAIPEFVGEMPALEVLQLW 379
Query: 530 NNHFTDSIPTEFGLLQRLEVLDLGGNELSGMIPNEVAELPKLRMLNLSRNKIEGSIPSLF 589
N+FT SIP G +L ++DL N+L+G +P + +L+ L N + G IP
Sbjct: 380 ENNFTGSIPQSLGKNGKLTLVDLSSNKLTGTLPPHMCSGNRLQTLIALGNFLFGPIPESL 559
Query: 590 R--SSLASLDLSGNRLNGKIPEILGFLGQLSMLNLSHNMLSGTIPSFSSMS--LDFVNIS 645
SL + + N LNG IP+ L L +L+ + N+LSG P S+S + + +S
Sbjct: 560 GKCESLTRIRMGQNFLNGSIPKGLFGLPKLTQVEFQDNLLSGEFPETGSVSHNIGQITLS 739
Query: 646 NNQLEGPLP 654
NN+L GPLP
Sbjct: 740 NNKLSGPLP 766
Score = 116 bits (290), Expect = 2e-26
Identities = 89/300 (29%), Positives = 130/300 (42%), Gaps = 3/300 (1%)
Frame = +2
Query: 310 LFLGM-NRLSGSIPATIGNLINLDSFSVQENNLTGTIPTTIGNLNRLTVFEVAANKLHGR 368
L+LG N G IP IGNL L F L+G IP +G L +L + N L G
Sbjct: 5 LYLGYYNNYQGGIPPEIGNLTQLLRFDAAYCGLSGEIPAELGKLQKLDTLFLQVNVLSGS 184
Query: 369 IPNGLYNITNWFSFIVSKNDFVGHLPSQICSGGLLTLLNADHNRFTGPIPTSLKNCSSIE 428
+ L ++ + S +S N G +P+ LTLLN NR G IP + ++E
Sbjct: 185 LTPELGHLKSLKSMDLSNNMLSGQVPASFAELKNLTLLNLFRNRLHGAIPEFVGEMPALE 364
Query: 429 RIRLEVNQIEGDIAQDFGVYPNLRYFDVSDNKLHGHISPNWGKSLNLDTFQISNNNISGV 488
++L N G I Q G L D+S NKL G + P+ S N + +
Sbjct: 365 VLQLWENNFTGSIPQSLGKNGKLTLVDLSSNKLTGTLPPH----------MCSGNRLQTL 514
Query: 489 IPLELIGLTKLGRLHLSSNQFTGKLPKELGGMKSLFDLKLSNNHFTDSIPTEFGLLQRLE 548
I L N G +P+ LG +SL +++ N SIP L +L
Sbjct: 515 IAL--------------GNFLFGPIPESLGKCESLTRIRMGQNFLNGSIPKGLFGLPKLT 652
Query: 549 VLDLGGNELSGMIPNEVAELPKLRMLNLSRNKIEGSIPSLFR--SSLASLDLSGNRLNGK 606
++ N LSG P + + + LS NK+ G +PS +S+ L L GN+ +G+
Sbjct: 653 QVEFQDNLLSGEFPETGSVSHNIGQITLSNNKLSGPLPSTIGNFTSMQKLLLDGNKFSGR 832
>BP033681
Length = 454
Score = 156 bits (395), Expect = 1e-38
Identities = 77/125 (61%), Positives = 98/125 (77%)
Frame = +1
Query: 1 MIMFIILFMISWPQAVAEDSEAQALLKWKHSFDNQSQSLLSTWKNTTNTCTKWKGIFCDN 60
M ++I+L ++ PQA AE+SEA ALLKWK S DNQSQ+LLSTW T + KW+GI CD
Sbjct: 88 MFLYILLTLLL-PQAAAEESEANALLKWKQSLDNQSQALLSTW--TGSDPCKWRGIQCDK 258
Query: 61 SKSISTINLENFGLKGTLHSLTFSSFSNLQTLNIYNNYFYGTIPPQIGNISKINTLNFSL 120
S SIS I+L N+GLKGTLH+L+FSSF NL+T++I+ N FYGTIPPQ+G++ K+N LNFS
Sbjct: 259 SSSISRISLANYGLKGTLHTLSFSSFPNLKTIDIFGNSFYGTIPPQVGDMPKVNILNFSE 438
Query: 121 NPIDG 125
N DG
Sbjct: 439 NSFDG 453
>AI967315
Length = 1308
Score = 155 bits (393), Expect = 2e-38
Identities = 100/295 (33%), Positives = 153/295 (50%), Gaps = 8/295 (2%)
Frame = +1
Query: 749 FENIIEATENFDDKYLIGVGSQGNVYKAELSSGMVVAVKKLHIITDEEISHFSSKSFMSE 808
+E + AT F + ++G G VYK L SG +AVK+L +E K F++E
Sbjct: 118 YEELFHATNGFSSENMVGKGGYAEVYKGRLESGDEIAVKRLTRTCRDERKE---KEFLTE 288
Query: 809 IETLSGIRHRNIIKLHGFCSHSKFSFLVYKFLEGGSLGQMLNSDTQATAFDWEKRVNVVK 868
I T+ + H N++ L G C + +LV++ GS+ +++ + A DW+ R +V
Sbjct: 289 IGTIGHVCHSNVMPLLGCCIDNGL-YLVFELSTVGSVASLIHDEKMAP-LDWKTRYKIVL 462
Query: 869 GVANALSYLHHDCSPPIIHRDISSKNVLLNLDYEAQVSDFGTAKFLKPGLLSWTQFA--- 925
G A L YLH C IIHRDI + N+LL D+E Q+SDFG AK+L WT +
Sbjct: 463 GTARGLHYLHKGCQRRIIHRDIKASNILLTEDFEPQISDFGLAKWLPS---QWTHHSIAP 633
Query: 926 --GTFGYAAPELAQTMEVNEKCDVYSFGVLALEIIVGKHPGDLISLFLSQSTRLMANNML 983
GTFG+ APE V+EK DV++FGV LE+I G+ P D L + + +
Sbjct: 634 IEGTFGHLAPEYYMHGVVDEKTDVFAFGVFLLEVISGRKPVDGSHQSLHTWAKPILSKWE 813
Query: 984 LIDVLDQRPQHVMKPVDEEVILIARLAFA---CLNQNPRSRPTMDQVSKMLAIGK 1035
+ ++D R + +V R+AFA C+ + RPTM +V +++ G+
Sbjct: 814 IEKLVDPRLEGCY-----DVTQFNRVAFAASLCIRASSTWRPTMSEVLEVMEEGE 963
>TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partial (77%)
Length = 1478
Score = 146 bits (368), Expect = 2e-35
Identities = 87/250 (34%), Positives = 133/250 (52%), Gaps = 8/250 (3%)
Frame = +1
Query: 724 EKNQTEEQTQRGVLFSIWSHDGKMM-----FENIIEATENFDDKYLIGVGSQGNVYKAEL 778
E+ + + +GV S S +GK F + +AT NF + LIG G G VYK L
Sbjct: 331 ERKRETKGKGKGVSSSNGSSNGKTAAASFGFRELADATRNFKEANLIGEGGFGKVYKGRL 510
Query: 779 SSGMVVAVKKLHIITDEEISHFSSKSFMSEIETLSGIRHRNIIKLHGFCSHSKFSFLVYK 838
++G VAVK+L + F+ E+ LS + H N+++L G+C+ LVY+
Sbjct: 511 TTGEAVAVKQL-----SHDGRQGFQEFVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYE 675
Query: 839 FLEGGSL-GQMLNSDTQATAFDWEKRVNVVKGVANALSYLHHDCSPPIIHRDISSKNVLL 897
++ GSL + +W R+ V G A L YLH PP+I+RD+ S N+LL
Sbjct: 676 YMPMGSLEDHLFELSHDKEPLNWSTRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILL 855
Query: 898 NLDYEAQVSDFGTAKFLKPGLLSW--TQFAGTFGYAAPELAQTMEVNEKCDVYSFGVLAL 955
+ ++ ++SDFG AK G + T+ GT+GY APE A + ++ K D+YSFGV+ L
Sbjct: 856 DNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLL 1035
Query: 956 EIIVGKHPGD 965
E++ G+ D
Sbjct: 1036ELLTGRRAID 1065
>TC17311 similar to UP|Q8K2Y2 (Q8K2Y2) Receptor-interacting serine-threonine
kinase 3, partial (7%)
Length = 606
Score = 145 bits (367), Expect = 2e-35
Identities = 77/165 (46%), Positives = 103/165 (61%), Gaps = 11/165 (6%)
Frame = +3
Query: 808 EIETLSGIRHRNIIKLHGFCSHSKFSFLVYKFLEGGSLGQMLNSDT---------QATAF 858
E+E LS IRH NI+KL S + LVY +LE SL + L+ + Q
Sbjct: 3 EVEILSNIRHNNIVKLQCCISSEESLLLVYDYLENLSLDRWLHKKSKPPTVSGSVQNNII 182
Query: 859 DWEKRVNVVKGVANALSYLHHDCSPPIIHRDISSKNVLLNLDYEAQVSDFGTAKF-LKP- 916
DW +R+++ GVA L Y+HHDCSPP++HRD+ N+LL+ + A+V+DFG A +KP
Sbjct: 183 DWPRRLHIAIGVAQGLCYMHHDCSPPVVHRDLKPSNILLDSQFNAKVADFGLAMMSVKPE 362
Query: 917 GLLSWTQFAGTFGYAAPELAQTMEVNEKCDVYSFGVLALEIIVGK 961
L + + AGTFGY APE A T+ VNEK DVYSFGV+ LE+ GK
Sbjct: 363 ELATMSAVAGTFGYIAPEYALTIRVNEKIDVYSFGVILLELTTGK 497
>TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete
Length = 2193
Score = 145 bits (366), Expect = 3e-35
Identities = 95/288 (32%), Positives = 149/288 (50%), Gaps = 12/288 (4%)
Frame = +1
Query: 749 FENIIEATENFDDKYLIGVGSQGNVYKAELSSGMVVAVKKLHIITDEEISHFSSKSFMSE 808
++ + +AT NF IG G G VY AEL G A+KK+ + E F+ E
Sbjct: 1057 YQELAKATNNFSLDNKIGQGGFGAVYYAELR-GKKTAIKKMDVQASTE--------FLCE 1209
Query: 809 IETLSGIRHRNIIKLHGFCSHSKFSFLVYKFLEGGSLGQMLNSDTQATAFDWEKRVNVVK 868
++ L+ + H N+++L G+C FLVY+ ++ G+LGQ L+ + W RV +
Sbjct: 1210 LKVLTHVHHLNLVRLIGYCVEGSL-FLVYEHIDNGNLGQYLHGSGKEP-LPWSSRVQIAL 1383
Query: 869 GVANALSYLHHDCSPPIIHRDISSKNVLLNLDYEAQVSDFGTAKFLKPGLLS-WTQFAGT 927
A L Y+H P IHRD+ S N+L++ + +V+DFG K ++ G + T+ GT
Sbjct: 1384 DAARGLEYIHEHTVPVYIHRDVKSANILIDKNLRGKVADFGLTKLIEVGNSTLQTRLVGT 1563
Query: 928 FGYAAPELAQTMEVNEKCDVYSFGVLALEIIVGKHPGDLISLFLSQSTRLMANNMLLIDV 987
FGY PE AQ +++ K DVY+FGV+ E+I K+ +++S L+A L +
Sbjct: 1564 FGYMPPEYAQYGDISPKIDVYAFGVVLFELISAKNAVLKTGELVAESKGLVA---LFEEA 1734
Query: 988 LDQRP--QHVMKPVD---------EEVILIARLAFACLNQNPRSRPTM 1024
L++ + K VD + V+ IA+L AC NP RP+M
Sbjct: 1735 LNKSDPCDALRKLVDPRLGENYPIDSVLKIAQLGRACTRDNPLLRPSM 1878
>TC9538 similar to UP|RLK5_ARATH (P47735) Receptor-like protein kinase 5
precursor , partial (10%)
Length = 1067
Score = 142 bits (357), Expect = 4e-34
Identities = 86/227 (37%), Positives = 124/227 (53%), Gaps = 14/227 (6%)
Frame = +2
Query: 819 NIIKLHGFCSHSKFSFLVYKFLEGGSLGQMLNSDT----------QATAFDWEKRVNVVK 868
NI++L S+ LVY++LE SL + L+ Q T DW KR+ +
Sbjct: 5 NIVRLLCCISNEASMLLVYEYLENHSLDKWLHLKPKSSSVSGVVQQYTVLDWPKRLKIAI 184
Query: 869 GVANALSYLHHDCSPPIIHRDISSKNVLLNLDYEAQVSDFGTAKFL-KPGLLS-WTQFAG 926
G A LSY+HHDCSPPI+HRD+ + N+LL+ + A+V+DFG A+ L KPG L+ + G
Sbjct: 185 GAAQGLSYMHHDCSPPIVHRDVKTSNILLDKQFNAKVADFGLARMLIKPGELNIMSTVIG 364
Query: 927 TFGYAAPELAQTMEVNEKCDVYSFGVLALEIIVGKHP--GDLISLFLSQSTRLMANNMLL 984
TFGY APE QT ++EK DVYSFGV+ LE+ GK GD S + R + +
Sbjct: 365 TFGYIAPEYVQTTRISEKVDVYSFGVVLLELTTGKEANYGDQHSSLAEWAWRHILIGSNV 544
Query: 985 IDVLDQRPQHVMKPVDEEVILIARLAFACLNQNPRSRPTMDQVSKML 1031
D+L + +E+ + +L C P +RP+M +V +L
Sbjct: 545 XDLLXKDVMEA--SYIDEMCSVFKLGVMCTATLPATRPSMKEVLPIL 679
>TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2, complete
Length = 2946
Score = 141 bits (356), Expect = 5e-34
Identities = 111/376 (29%), Positives = 179/376 (47%), Gaps = 24/376 (6%)
Frame = +1
Query: 680 LDPCGSRKSKNV--LRSVLIALGALILVLFGVGISMYTLGRRKKSNEKNQTE-------- 729
LD ++KS N L L+ + A ++ + +G+++ T +RKK+ ++ E
Sbjct: 1417 LDRTRNKKSINTKKLAGSLVVIIAFVIFITILGLAISTCIQRKKNKRGDEGEIGIINHWK 1596
Query: 730 -----EQTQRGVLFSIWSHDGKMMFENIIEATENFDDKYLIGVGSQGNVYKAELSSGMVV 784
E +F F I AT +F +G G G VYK L++G +
Sbjct: 1597 DKRGDEDIDLATIFD---------FSTISSATNHFSLSNKLGEGGFGPVYKGLLANGQEI 1749
Query: 785 AVKKLHIITDEEISHFSSKSFMSEIETLSGIRHRNIIKLHGFCSHSKFSFLVYKFLEGGS 844
AVK+L S + F +EI+ ++ ++HRN++KL G H + K ++
Sbjct: 1750 AVKRL-----SNTSGQGMEEFKNEIKLIARLQHRNLVKLFGCSVHQDENSHANKKMK--- 1905
Query: 845 LGQMLNSDTQATAFDWEKRVNVVKGVANALSYLHHDCSPPIIHRDISSKNVLLNLDYEAQ 904
+L T++ DW KR+ ++ G+A L YLH D IIHRD+ + N+LL+ + +
Sbjct: 1906 ---ILLDSTRSKLVDWNKRLQIIDGIARGLLYLHQDSRLRIIHRDLKTSNILLDDEMNPK 2076
Query: 905 VSDFGTAK-FLKPGLLSWT-QFAGTFGYAAPELAQTMEVNEKCDVYSFGVLALEIIVGKH 962
+SDFG A+ F+ + + T + GT+GY PE A + K DV+SFGV+ LEII GK
Sbjct: 2077 ISDFGLARIFIGDQVEARTKRVMGTYGYMPPEYAVHGSFSIKSDVFSFGVIVLEIISGKK 2256
Query: 963 PGDLIS-----LFLSQSTRLMANN--MLLIDVLDQRPQHVMKPVDEEVILIARLAFACLN 1015
G LS + RL + L+D L P + E++ +A C+
Sbjct: 2257 IGRFYDPHHHLNLLSHAWRLWIEERPLELVDELLDDP-----VIPTEILRYIHVALLCVQ 2421
Query: 1016 QNPRSRPTMDQVSKML 1031
+ P +RP M + ML
Sbjct: 2422 RRPENRPDMLSIVLML 2469
>CN825263
Length = 663
Score = 140 bits (353), Expect = 1e-33
Identities = 79/199 (39%), Positives = 111/199 (55%), Gaps = 3/199 (1%)
Frame = +1
Query: 771 GNVYKAELSSGMVVAVKKLHIITDEEISHFSSKSFMSEIETLSGIRHRNIIKLHGFCSHS 830
G VYK L+ G VAVK L + + F++E+E LS + HRN++KL G C
Sbjct: 7 GLVYKGILNDGRDVAVKIL-----KRDDQRGGREFLAEVEMLSRLHHRNLVKLIGICIEK 171
Query: 831 KFSFLVYKFLEGGSLGQMLN-SDTQATAFDWEKRVNVVKGVANALSYLHHDCSPPIIHRD 889
+ L+Y+ + GS+ L+ +D + DW R+ + G A L+YLH D +P +IHRD
Sbjct: 172 QTRCLIYELVPNGSVESHLHGADKETGPLDWNARMKIALGAARGLAYLHEDSNPCVIHRD 351
Query: 890 ISSKNVLLNLDYEAQVSDFGTAKFL--KPGLLSWTQFAGTFGYAAPELAQTMEVNEKCDV 947
S N+LL D+ +VSDFG A+ + T GTFGY APE A T + K DV
Sbjct: 352 FKSSNILLECDFTPKVSDFGLARTALDEGNKHISTHVMGTFGYLAPEYAMTGHLLVKSDV 531
Query: 948 YSFGVLALEIIVGKHPGDL 966
YS+GV+ LE++ G P DL
Sbjct: 532 YSYGVVLLELLTGTKPVDL 588
>TC15149 similar to UP|Q9LVI6 (Q9LVI6) Probable receptor-like protein kinase
protein (AT3g17840/MEB5_6), partial (45%)
Length = 1489
Score = 140 bits (352), Expect = 1e-33
Identities = 89/274 (32%), Positives = 141/274 (50%), Gaps = 6/274 (2%)
Frame = +3
Query: 764 LIGVGSQGNVYKAELSSGMVVAVKKLHIITDEEISHFSSKSFMSEIETLSGIRHRNIIKL 823
++G G+ G YKA L +G+VVAVK+L +T S K F +IET+ + H++++ L
Sbjct: 291 VLGKGTFGTAYKAVLETGLVVAVKRLKDVT------ISEKEFKDKIETVGAMDHQSLVPL 452
Query: 824 HGFCSHSKFSFLVYKFLEGGSLGQMLNSDTQA--TAFDWEKRVNVVKGVANALSYLHHDC 881
+ LVY ++ GSL +L+ + A T +WE R + G A + YLH
Sbjct: 453 RAYYFSRDEKLLVYDYMPMGSLSALLHGNKGAGRTPLNWEIRSGIALGAARGIEYLHSQ- 629
Query: 882 SPPIIHRDISSKNVLLNLDYEAQVSDFGTAKFLKPGLLSWTQFAGTFGYAAPELAQTMEV 941
P + H +I + N+LL YEA+VSDFG A + P GY APE+ +V
Sbjct: 630 GPNVSHGNIKASNILLTKSYEAKVSDFGLAHLVGPSSTP----NRVAGYRAPEVTDPRKV 797
Query: 942 NEKCDVYSFGVLALEIIVGKHPGDLI----SLFLSQSTRLMANNMLLIDVLDQRPQHVMK 997
++K DVYSFGVL LE++ GK P + + L + + + +V D
Sbjct: 798 SQKADVYSFGVLLLELLTGKAPTHALLNEEGVDLPRWVQSLVREEWTSEVFDLELLRYQN 977
Query: 998 PVDEEVILIARLAFACLNQNPRSRPTMDQVSKML 1031
V+EE++ + +LA C P RP++ +V++ +
Sbjct: 978 -VEEEMVQLLQLAVDCAAPYPDRRPSIAEVTRSI 1076
>AV416024
Length = 249
Score = 137 bits (346), Expect = 7e-33
Identities = 66/82 (80%), Positives = 75/82 (90%)
Frame = +2
Query: 759 FDDKYLIGVGSQGNVYKAELSSGMVVAVKKLHIITDEEISHFSSKSFMSEIETLSGIRHR 818
FDDKYLIGVG QGNVY AEL +G+VVAVKKLH +TDEE+S+FSSK+F SEI+TL+ IRHR
Sbjct: 2 FDDKYLIGVGGQGNVYMAELPAGLVVAVKKLHSVTDEEMSNFSSKAFNSEIQTLTEIRHR 181
Query: 819 NIIKLHGFCSHSKFSFLVYKFL 840
NIIKLHGFC HSKFSFLVY+FL
Sbjct: 182 NIIKLHGFCLHSKFSFLVYEFL 247
>AW719637
Length = 571
Score = 132 bits (332), Expect = 3e-31
Identities = 75/186 (40%), Positives = 108/186 (57%)
Frame = +2
Query: 176 IPPEIGKLNKLWFLSIQKCNLIGSIPKEIGFLTNLTLIDLSNNILSGVIPETIGNMSKLN 235
IP EI L+ L LS+ L G+IP IG L++L + L +N LSG IP++IG++ L
Sbjct: 14 IPEEICSLSNLQSLSLHTNFLEGNIPSNIGNLSSLVNLTLFDNHLSGEIPKSIGSLRNLQ 193
Query: 236 KLYLAKNTKLYGPIPHSLWNMSSLTLIYLFNMSLSGSIPESVENLINVNELALDRNRLSG 295
N L G IP + N +SL ++ L S+SGS+P S++ L + +A+ LSG
Sbjct: 194 VFRAGGNKNLKGEIPWEIGNCTSLVMLGLAETSISGSLPSSIQLLKRIKTIAIYTTLLSG 373
Query: 296 TIPSTIGNLKNLQYLFLGMNRLSGSIPATIGNLINLDSFSVQENNLTGTIPTTIGNLNRL 355
+IP IGN LQ L+L N +SGSIP+ IG L L S + +NN+ GTIP IG +
Sbjct: 374 SIPEEIGNCSELQNLYLYQNSISGSIPSQIGELSKLKSLLLWQNNIVGTIPEEIGRCTEM 553
Query: 356 TVFEVA 361
V +++
Sbjct: 554 EVIDLS 571
Score = 115 bits (289), Expect = 3e-26
Identities = 73/212 (34%), Positives = 104/212 (48%)
Frame = +2
Query: 101 GTIPPQIGNISKINTLNFSLNPIDGSIPQEMFTLKSLQNIDFSFCKLSGAIPNSIGNLSN 160
G IP +I ++S + +L+ N ++G+IP + L SL N+ LSG IP SIG+L N
Sbjct: 8 GEIPEEICSLSNLQSLSLHTNFLEGNIPSNIGNLSSLVNLTLFDNHLSGEIPKSIGSLRN 187
Query: 161 LLYLDLGGNNFVGTPIPPEIGKLNKLWFLSIQKCNLIGSIPKEIGFLTNLTLIDLSNNIL 220
L GGN + IP EIG L L + + ++ GS+P I L + I + +L
Sbjct: 188 LQVFRAGGNKNLKGEIPWEIGNCTSLVMLGLAETSISGSLPSSIQLLKRIKTIAIYTTLL 367
Query: 221 SGVIPETIGNMSKLNKLYLAKNTKLYGPIPHSLWNMSSLTLIYLFNMSLSGSIPESVENL 280
SG IPE IGN S+L LYL +N S+SGSIP + L
Sbjct: 368 SGSIPEEIGNCSELQNLYLYQN-------------------------SISGSIPSQIGEL 472
Query: 281 INVNELALDRNRLSGTIPSTIGNLKNLQYLFL 312
+ L L +N + GTIP IG ++ + L
Sbjct: 473 SKLKSLLLWQNNIVGTIPEEIGRCTEMEVIDL 568
Score = 107 bits (268), Expect = 7e-24
Identities = 67/183 (36%), Positives = 102/183 (55%), Gaps = 1/183 (0%)
Frame = +2
Query: 196 LIGSIPKEIGFLTNLTLIDLSNNILSGVIPETIGNMSKLNKLYLAKNTKLYGPIPHSLWN 255
L+G IP+EI L+NL + L N L G IP IGN+S L L L N L G IP S+ +
Sbjct: 2 LLGEIPEEICSLSNLQSLSLHTNFLEGNIPSNIGNLSSLVNLTLFDN-HLSGEIPKSIGS 178
Query: 256 MSSLTLIYLF-NMSLSGSIPESVENLINVNELALDRNRLSGTIPSTIGNLKNLQYLFLGM 314
+ +L + N +L G IP + N ++ L L +SG++PS+I LK ++ + +
Sbjct: 179 LRNLQVFRAGGNKNLKGEIPWEIGNCTSLVMLGLAETSISGSLPSSIQLLKRIKTIAIYT 358
Query: 315 NRLSGSIPATIGNLINLDSFSVQENNLTGTIPTTIGNLNRLTVFEVAANKLHGRIPNGLY 374
LSGSIP IGN L + + +N+++G+IP+ IG L++L + N + G IP +
Sbjct: 359 TLLSGSIPEEIGNCSELQNLYLYQNSISGSIPSQIGELSKLKSLLLWQNNIVGTIPEEIG 538
Query: 375 NIT 377
T
Sbjct: 539 RCT 547
Score = 100 bits (248), Expect = 2e-21
Identities = 64/179 (35%), Positives = 100/179 (55%), Gaps = 1/179 (0%)
Frame = +2
Query: 220 LSGVIPETIGNMSKLNKLYLAKNTKLYGPIPHSLWNMSSLTLIYLFNMSLSGSIPESVEN 279
L G IPE I ++S L L L N L G IP ++ N+SSL + LF+ LSG IP+S+ +
Sbjct: 2 LLGEIPEEICSLSNLQSLSLHTNF-LEGNIPSNIGNLSSLVNLTLFDNHLSGEIPKSIGS 178
Query: 280 LINVNELALDRNR-LSGTIPSTIGNLKNLQYLFLGMNRLSGSIPATIGNLINLDSFSVQE 338
L N+ N+ L G IP IGN +L L L +SGS+P++I L + + ++
Sbjct: 179 LRNLQVFRAGGNKNLKGEIPWEIGNCTSLVMLGLAETSISGSLPSSIQLLKRIKTIAIYT 358
Query: 339 NNLTGTIPTTIGNLNRLTVFEVAANKLHGRIPNGLYNITNWFSFIVSKNDFVGHLPSQI 397
L+G+IP IGN + L + N + G IP+ + ++ S ++ +N+ VG +P +I
Sbjct: 359 TLLSGSIPEEIGNCSELQNLYLYQNSISGSIPSQIGELSKLKSLLLWQNNIVGTIPEEI 535
Score = 95.5 bits (236), Expect = 4e-20
Identities = 57/189 (30%), Positives = 98/189 (51%), Gaps = 1/189 (0%)
Frame = +2
Query: 390 VGHLPSQICSGGLLTLLNADHNRFTGPIPTSLKNCSSIERIRLEVNQIEGDIAQDFGVYP 449
+G +P +ICS L L+ N G IP+++ N SS+ + L N + G+I + G
Sbjct: 5 LGEIPEEICSLSNLQSLSLHTNFLEGNIPSNIGNLSSLVNLTLFDNHLSGEIPKSIGSLR 184
Query: 450 NLRYFDVSDNK-LHGHISPNWGKSLNLDTFQISNNNISGVIPLELIGLTKLGRLHLSSNQ 508
NL+ F NK L G I G +L ++ +ISG +P + L ++ + + +
Sbjct: 185 NLQVFRAGGNKNLKGEIPWEIGNCTSLVMLGLAETSISGSLPSSIQLLKRIKTIAIYTTL 364
Query: 509 FTGKLPKELGGMKSLFDLKLSNNHFTDSIPTEFGLLQRLEVLDLGGNELSGMIPNEVAEL 568
+G +P+E+G L +L L N + SIP++ G L +L+ L L N + G IP E+
Sbjct: 365 LSGSIPEEIGNCSELQNLYLYQNSISGSIPSQIGELSKLKSLLLWQNNIVGTIPEEIGRC 544
Query: 569 PKLRMLNLS 577
++ +++LS
Sbjct: 545 TEMEVIDLS 571
Score = 95.5 bits (236), Expect = 4e-20
Identities = 61/174 (35%), Positives = 92/174 (52%), Gaps = 5/174 (2%)
Frame = +2
Query: 487 GVIPLELIGLTKLGRLHLSSNQFTGKLPKELGGMKSLFDLKLSNNHFTDSIPTEFGLLQR 546
G IP E+ L+ L L L +N G +P +G + SL +L L +NH + IP G L+
Sbjct: 8 GEIPEEICSLSNLQSLSLHTNFLEGNIPSNIGNLSSLVNLTLFDNHLSGEIPKSIGSLRN 187
Query: 547 LEVLDLGGNE-LSGMIPNEVAELPKLRMLNLSRNKIEGSIPSLFR--SSLASLDLSGNRL 603
L+V GGN+ L G IP E+ L ML L+ I GS+PS + + ++ + L
Sbjct: 188 LQVFRAGGNKNLKGEIPWEIGNCTSLVMLGLAETSISGSLPSSIQLLKRIKTIAIYTTLL 367
Query: 604 NGKIPEILGFLGQLSMLNLSHNMLSGTIPS-FSSMS-LDFVNISNNQLEGPLPD 655
+G IPE +G +L L L N +SG+IPS +S L + + N + G +P+
Sbjct: 368 SGSIPEEIGNCSELQNLYLYQNSISGSIPSQIGELSKLKSLLLWQNNIVGTIPE 529
Score = 90.5 bits (223), Expect = 1e-18
Identities = 61/188 (32%), Positives = 97/188 (51%), Gaps = 3/188 (1%)
Frame = +2
Query: 439 GDIAQDFGVYPNLRYFDVSDNKLHGHISPNWGKSLNLDTFQISNNNISGVIPLELIGLTK 498
G+I ++ NL+ + N L G+I N G +L + +N++SG IP + L
Sbjct: 8 GEIPEEICSLSNLQSLSLHTNFLEGNIPSNIGNLSSLVNLTLFDNHLSGEIPKSIGSLRN 187
Query: 499 LGRLHLSSNQ-FTGKLPKELGGMKSLFDLKLSNNHFTDSIPTEFGLLQRLEVLDLGGNEL 557
L N+ G++P E+G SL L L+ + S+P+ LL+R++ + + L
Sbjct: 188 LQVFRAGGNKNLKGEIPWEIGNCTSLVMLGLAETSISGSLPSSIQLLKRIKTIAIYTTLL 367
Query: 558 SGMIPNEVAELPKLRMLNLSRNKIEGSIPSLF--RSSLASLDLSGNRLNGKIPEILGFLG 615
SG IP E+ +L+ L L +N I GSIPS S L SL L N + G IPE +G
Sbjct: 368 SGSIPEEIGNCSELQNLYLYQNSISGSIPSQIGELSKLKSLLLWQNNIVGTIPEEIGRCT 547
Query: 616 QLSMLNLS 623
++ +++LS
Sbjct: 548 EMEVIDLS 571
Score = 84.3 bits (207), Expect = 9e-17
Identities = 58/158 (36%), Positives = 85/158 (53%), Gaps = 1/158 (0%)
Frame = +2
Query: 60 NSKSISTINLENFGLKGTLHSLTFSSFSNLQTLNIYNNY-FYGTIPPQIGNISKINTLNF 118
N S+ + L + L G + + S NLQ N G IP +IGN + + L
Sbjct: 104 NLSSLVNLTLFDNHLSGEIPK-SIGSLRNLQVFRAGGNKNLKGEIPWEIGNCTSLVMLGL 280
Query: 119 SLNPIDGSIPQEMFTLKSLQNIDFSFCKLSGAIPNSIGNLSNLLYLDLGGNNFVGTPIPP 178
+ I GS+P + LK ++ I LSG+IP IGN S L L L N+ G+ IP
Sbjct: 281 AETSISGSLPSSIQLLKRIKTIAIYTTLLSGSIPEEIGNCSELQNLYLYQNSISGS-IPS 457
Query: 179 EIGKLNKLWFLSIQKCNLIGSIPKEIGFLTNLTLIDLS 216
+IG+L+KL L + + N++G+IP+EIG T + +IDLS
Sbjct: 458 QIGELSKLKSLLLWQNNIVGTIPEEIGRCTEMEVIDLS 571
Score = 54.7 bits (130), Expect = 7e-08
Identities = 38/105 (36%), Positives = 55/105 (52%), Gaps = 5/105 (4%)
Frame = +2
Query: 557 LSGMIPNEVAELPKLRMLNLSRNKIEGSIPSLF--RSSLASLDLSGNRLNGKIPEILGFL 614
L G IP E+ L L+ L+L N +EG+IPS SSL +L L N L+G+IP+ +G L
Sbjct: 2 LLGEIPEEICSLSNLQSLSLHTNFLEGNIPSNIGNLSSLVNLTLFDNHLSGEIPKSIGSL 181
Query: 615 GQLSMLNLSHNM-LSGTIP--SFSSMSLDFVNISNNQLEGPLPDN 656
L + N L G IP + SL + ++ + G LP +
Sbjct: 182 RNLQVFRAGGNKNLKGEIPWEIGNCTSLVMLGLAETSISGSLPSS 316
>TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-like protein,
partial (46%)
Length = 941
Score = 129 bits (325), Expect = 2e-30
Identities = 93/270 (34%), Positives = 139/270 (51%), Gaps = 6/270 (2%)
Frame = +3
Query: 764 LIGVGSQGNVYKAELSSGMVVAVKKLHIITDEEISHFSSKSFMSEIETLSGIRHRNIIKL 823
L+GVG G VY E+ G VA+K+ + ++++ + F + EIE LS +RHR+++ L
Sbjct: 12 LLGVGGFGKVYYGEVDGGTKVAIKRGNPLSEQGVHEFQT-----EIEMLSKLRHRHLVSL 176
Query: 824 HGFCSHSKFSFLVYKFLEGGSLGQMLNSDTQATAFDWEKRVNVVKGVANALSYLHHDCSP 883
G+C + LVY + G+L + L TQ W++R+ + G A L YLH
Sbjct: 177 IGYCEENTEMILVYDHMAYGTLREHLYK-TQKPPLPWKQRLEICIGAARGLHYLHTGAKY 353
Query: 884 PIIHRDISSKNVLLNLDYEAQVSDFGTAKFLKPGLLSW---TQFAGTFGYAAPELAQTME 940
IIHRD+ + N+LL+ + A+VSDFG +K P L + T G+FGY PE + +
Sbjct: 354 TIIHRDVKTTNILLDEKWVAKVSDFGLSK-TGPTLDNTHVSTVVKGSFGYLDPEYFRRQQ 530
Query: 941 VNEKCDVYSFGVLALEIIVGKHPGDLISLFLSQ-STRLMANNMLLIDVLDQ--RPQHVMK 997
+ +K DVYSFGV+ EI+ + P SL Q S A++ +LDQ P K
Sbjct: 531 LTDKSDVYSFGVVLFEILCAR-PALNPSLAKEQVSLAEWASHCYNKGILDQILDPYLKGK 707
Query: 998 PVDEEVILIARLAFACLNQNPRSRPTMDQV 1027
E A A C++ RP+M V
Sbjct: 708 IAPECFKKFAETAMKCVSDQGIERPSMGDV 797
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.137 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,851,161
Number of Sequences: 28460
Number of extensions: 249473
Number of successful extensions: 2637
Number of sequences better than 10.0: 378
Number of HSP's better than 10.0 without gapping: 1579
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2087
length of query: 1052
length of database: 4,897,600
effective HSP length: 100
effective length of query: 952
effective length of database: 2,051,600
effective search space: 1953123200
effective search space used: 1953123200
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC133779.2


