

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133779.10 + phase: 2 /pseudo
(609 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
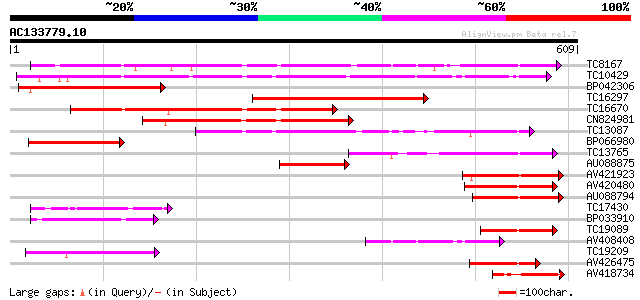
Score E
Sequences producing significant alignments: (bits) Value
TC8167 similar to PIR|JC7519|JC7519 subtilisin-like serine prote... 378 e-105
TC10429 similar to UP|Q9ZR46 (Q9ZR46) P69C protein, partial (52%) 337 3e-93
BP042306 273 6e-74
TC16297 weakly similar to UP|Q93WQ0 (Q93WQ0) Subtilisin-type pro... 256 6e-69
TC16670 similar to PIR|JC7519|JC7519 subtilisin-like serine prot... 237 5e-63
CN824981 216 8e-57
TC13087 similar to UP|Q9FIF8 (Q9FIF8) Serine protease-like prote... 209 1e-54
BP066980 174 4e-44
TC13765 similar to UP|Q9LLL8 (Q9LLL8) Subtilisin-type serine end... 131 3e-31
AU088875 119 1e-27
AV421923 119 1e-27
AV420480 117 5e-27
AU088794 112 2e-25
TC17430 weakly similar to UP|Q9ZTT3 (Q9ZTT3) Subtilisin-like pro... 102 1e-22
BP033910 100 7e-22
TC19089 similar to UP|Q9ZTT3 (Q9ZTT3) Subtilisin-like protease C... 100 9e-22
AV408408 95 4e-20
TC19209 similar to GB|BAB11244.1|10177874|AB010074 serine protea... 92 3e-19
AV426475 88 3e-18
AV418734 82 2e-16
>TC8167 similar to PIR|JC7519|JC7519 subtilisin-like serine proteinase -
Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (65%)
Length = 2760
Score = 378 bits (971), Expect = e-105
Identities = 235/594 (39%), Positives = 329/594 (54%), Gaps = 24/594 (4%)
Frame = +3
Query: 23 YIVYMGAHSHGPT-PTSVDLETATSSHYDLLGSIVGSKEEAKEAIIYSYNKQINGFAAML 81
YIV+M H+ PT D TA+ S+ E + + ++Y+Y+ NGFAA L
Sbjct: 150 YIVHMNHHTKPQIYPTRRDWYTASLRSL----SLTTDSETSDDPLLYAYDTAYNGFAASL 317
Query: 82 EEEEAAQLAKNPKVVSVFLSKEHKLHTTRSWEFLGLHGNDINSAWQKGRFGE------NT 135
+E++A L + V+ ++ + LHTTR+ +FLGL W+ R E +
Sbjct: 318 DEQQAQTLLGSDSVLGLYEDTLYHLHTTRTPQFLGLDTQ--TGLWEGHRTLELDQASRDV 491
Query: 136 IIANIDTGVWPESRSFSDRGIGPIPAKWRGGSSKKVP-----CNRKLIGARFFSDAYERY 190
II +DTGVWPES SF+D G+ IP++WRG CNRKLIGAR FS +
Sbjct: 492 IIGVLDTGVWPESPSFNDAGMPEIPSRWRGECENATDFSSSLCNRKLIGARSFSRGFHMA 671
Query: 191 NGK-----LPTSQRTARDFVGHGTHTLSTAGGNFVPGASIFNIGNGTIKGGSPRARVATY 245
G + RD GHGTHT STA G+ V AS+ +GT +G +P+ARVATY
Sbjct: 672 AGNDGGFGKEREPPSPRDSDGHGTHTASTAAGSHVGNASLLGYASGTARGMAPQARVATY 851
Query: 246 KVCWSLTDAASCFGADVLSAIDQAIDDGVDIISVSAGGPSSTNSEEIFTDEVSIGAFHAL 305
KVCWS CF +D+L+ +D+AI DGVD++S+S GG S F D ++IGAF A+
Sbjct: 852 KVCWS----DGCFASDILAGMDRAIRDGVDVLSLSLGGGSGP----YFRDTIAIGAFAAM 1007
Query: 306 ARNILLVASAGNEGPTPGSVVNVAPWVFTVAASTIDRDFSSTITIGDQI-IRGASLFVDL 364
R I + SAGN GP+ S+ NVAPW+ TV A T+DRDF ++ +G++ G SL+
Sbjct: 1008ERGIFVSCSAGNSGPSKASLANVAPWLMTVGAGTLDRDFPASALLGNKKRFAGVSLYSGK 1187
Query: 365 PPNQSFTLVNSIDAKFSNATTRDARFCRPRTLDPSKVKGKIVACAREGKIKSVAEGQEAL 424
+ +S + + C P +LDP+ V+GK+V C R G V +G+
Sbjct: 1188GMG-----AEPVGLVYSKGSNQSGILCLPGSLDPAVVRGKVVLCDR-GLNARVEKGKVVK 1349
Query: 425 SAGAKGMFLENQPKVSGNTLLSEPHVLSTV------GGNGQAAITAPPRLGVTATDTIES 478
AG GM L N +G L+++ H+L V G + +T+ P TA
Sbjct: 1350EAGGIGMILTNTA-ANGEELVADSHLLPAVAVGRIVGDQIREYVTSDPN--PTAV----- 1505
Query: 479 GTKIRFSQAITLIGRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASASN 538
S + T++ +P+PV+A+FSSRGPN + ILKPDV PGVNILA +S S
Sbjct: 1506-----LSFSGTVLNVRPSPVVAAFSSRGPNMITKQILKPDVIGPGVNILAGWSEAIGPSG 1670
Query: 539 LLTDNRRGFPFNVMQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTVITY 592
L D+R+ FN+M GTSMSCPH++G L+K HP+WSP+AIKSA+MTT +
Sbjct: 1671LPQDSRKS-QFNIMSGTSMSCPHISGLGALLKAAHPDWSPSAIKSALMTTAYVH 1829
>TC10429 similar to UP|Q9ZR46 (Q9ZR46) P69C protein, partial (52%)
Length = 1742
Score = 337 bits (864), Expect = 3e-93
Identities = 228/603 (37%), Positives = 312/603 (50%), Gaps = 28/603 (4%)
Frame = +3
Query: 8 KLLVFYYYFFFLFQCYIVYMGAH----SHGPTPTSVDLETATSSHYDLL-----GSIVGS 58
K+L+ YF F+ +++ H S PT T+ ET++S Y + G ++
Sbjct: 18 KILLNMDYFLFIALTFLLTFHVHNAQGSELPTTTTESTETSSSKIYIIHVTGPEGKMLTE 197
Query: 59 KE-----------------EAKEAIIYSYNKQINGFAAMLEEEEAAQLAKNPKVVSVFLS 101
E E + +IYSY + GFAA L +EE + + + + + L
Sbjct: 198 SEDLESWYHSFLPPTLKSSEEQPRVIYSYKNVLRGFAASLTQEELSAVERKMALFQLILR 377
Query: 102 KEHKLHTTRSWEFLGLHGNDINSAWQKGRFGENTIIANIDTGVWPESRSFSDRGIGPIPA 161
+ + +FLGL + W++ FG+ II +D+G+ P SFSD GI P P
Sbjct: 378 GCYIVKPLIPPKFLGLQQD--TGVWKESNFGKGVIIGVLDSGITPGHPSFSDVGIPPPPP 551
Query: 162 KWRGGSSKKVP-CNRKLIGARFFSDAYERYNGKLPTSQRTARDFVGHGTHTLSTAGGNFV 220
KW+G V CN KLIGAR F+ A E NGK D GHGTHT STA G FV
Sbjct: 552 KWKGRCDLNVTACNNKLIGARAFNLAAEAMNGK---KAEAPIDEDGHGTHTASTAAGAFV 722
Query: 221 PGASIFNIGNGTIKGGSPRARVATYKVCWSLTDAASCFGADVLSAIDQAIDDGVDIISVS 280
A + GT G +P A +A YKVC+ C +D+L+A+D A++DGVD+IS+S
Sbjct: 723 NYAEVLGNAKGTAAGMAPHAHLAIYKVCFG----EDCPESDILAALDAAVEDGVDVISIS 890
Query: 281 AGGPSSTNSEEIFTDEVSIGAFHALARNILLVASAGNEGPTPGSVVNVAPWVFTVAASTI 340
G + F D +IGAF A+ + I + +AGN GP S+VN APW+ TV ASTI
Sbjct: 891 LG---LSEPPPFFNDSTAIGAFAAMQKGIFVSCAAGNSGPFNSSIVNAAPWILTVGASTI 1061
Query: 341 DRDFSSTITIGD-QIIRGASLFVDLPPNQSFTLVNSIDAKFSNATTRDARFCRPRTLDPS 399
DR +T +G+ Q G S+F SFT A ++ FC +LD S
Sbjct: 1062DRRIVATAKLGNGQEFDGESVF----QPSSFTPTLLPLAYAGKNGKEESAFCANGSLDDS 1229
Query: 400 KVKGKIVACAREGKIKSVAEGQEALSAGAKGMFLENQPKVSGNTLLSEPHVLSTVGGNGQ 459
+GK+V C R G I +A+G+E AG M L N + + +L ++ H L +
Sbjct: 1230AFRGKVVLCERGGGIARIAKGEEVKRAGGAAMILMND-ETNAFSLSADVHALPATHVSYA 1406
Query: 460 AAITAPPRLGVTATDTIESGTKIRFSQAITLIGRKPAPVMASFSSRGPNQVQPYILKPDV 519
A I + TAT T I F T+IG AP +ASFSSRGPN P ILKPD+
Sbjct: 1407AGIEIKAYINSTATPT----ATILFKG--TVIGNSLAPAVASFSSRGPNLPSPGILKPDI 1568
Query: 520 TAPGVNILAAYSLFASASNLLTDNRRGFPFNVMQGTSMSCPHVAGTAGLIKTLHPNWSPA 579
PGVNILAA+ S S TD++ FN+ GTSMSCPH++G A L+K+ HP+WSPA
Sbjct: 1569IGPGVNILAAWPFPLSNS---TDSK--LTFNIESGTSMSCPHLSGIAALLKSSHPHWSPA 1733
Query: 580 AIK 582
AIK
Sbjct: 1734AIK 1742
>BP042306
Length = 549
Score = 273 bits (698), Expect = 6e-74
Identities = 131/169 (77%), Positives = 148/169 (87%), Gaps = 11/169 (6%)
Frame = +3
Query: 10 LVFYYYFFFLF-----------QCYIVYMGAHSHGPTPTSVDLETATSSHYDLLGSIVGS 58
L+F +F LF +CYIVY+GAHSHGPTP+SVDLETATSSHYDLLGS++GS
Sbjct: 39 LLFLVSYFLLFISLLNSVHASKKCYIVYLGAHSHGPTPSSVDLETATSSHYDLLGSVLGS 218
Query: 59 KEEAKEAIIYSYNKQINGFAAMLEEEEAAQLAKNPKVVSVFLSKEHKLHTTRSWEFLGLH 118
E+AKEAI+YSYNK INGFAA+LEEEEAA++AKNPKVVSVF+SKEHKLHTTRSWEFLGL
Sbjct: 219 HEKAKEAILYSYNKHINGFAALLEEEEAARIAKNPKVVSVFVSKEHKLHTTRSWEFLGLR 398
Query: 119 GNDINSAWQKGRFGENTIIANIDTGVWPESRSFSDRGIGPIPAKWRGGS 167
GN NSAWQKGRFGENTIIANIDTGVWPES+SFSD+GIG +PAKWRGG+
Sbjct: 399 GNGGNSAWQKGRFGENTIIANIDTGVWPESQSFSDKGIGSVPAKWRGGN 545
>TC16297 weakly similar to UP|Q93WQ0 (Q93WQ0) Subtilisin-type protease,
partial (10%)
Length = 574
Score = 256 bits (655), Expect = 6e-69
Identities = 135/191 (70%), Positives = 156/191 (80%), Gaps = 1/191 (0%)
Frame = +2
Query: 261 DVLSAIDQAIDDGVDIISVSAGGPSSTNSEEIFTDEVSIGAFHALARNILLVASAGNEGP 320
DVL+AIDQAI DGVD+ SVSAGG S ++EEIFTDEVSIG+ HALA+NIL+VASAGN+GP
Sbjct: 2 DVLAAIDQAITDGVDVNSVSAGGRSIISAEEIFTDEVSIGSVHALAKNILVVASAGNDGP 181
Query: 321 TPGSVVNVAPWVFTVAASTIDRDFSSTITIG-DQIIRGASLFVDLPPNQSFTLVNSIDAK 379
G+V+NVAPWVFTVAASTIDR+FSSTIT+G +Q I GASLFV+LPPNQ F+L+ S DAK
Sbjct: 182 ALGTVLNVAPWVFTVAASTIDREFSSTITLGNNQQITGASLFVNLPPNQLFSLILSTDAK 361
Query: 380 FSNATTRDARFCRPRTLDPSKVKGKIVACAREGKIKSVAEGQEALSAGAKGMFLENQPKV 439
+NAT RDA+ CRP TLDP+KVKGK+V C R GKIKSVAEG EALS+GAKGM LE
Sbjct: 362 LANATNRDAQLCRPGTLDPAKVKGKVVVCTRGGKIKSVAEGSEALSSGAKGMVLE*SKAK 541
Query: 440 SGNTLLSEPHV 450
EPHV
Sbjct: 542 WKTHFFLEPHV 574
>TC16670 similar to PIR|JC7519|JC7519 subtilisin-like serine proteinase -
Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (42%)
Length = 1183
Score = 237 bits (604), Expect = 5e-63
Identities = 127/294 (43%), Positives = 180/294 (61%), Gaps = 7/294 (2%)
Frame = +2
Query: 66 IIYSYNKQINGFAAMLEEEEAAQLAKNPKVVSVFLSKEHKLHTTRSWEFLGLHGNDINSA 125
++Y+Y+ I+GF+ L EEA L V++V ++ +LHTTR+ FLGL +
Sbjct: 332 MLYTYDNAIHGFSTRLTPEEARLLEAQTGVLAVLPERKFELHTTRTPLFLGLDKSA--DM 505
Query: 126 WQKGRFGENTIIANIDTGVWPESRSFSDRGIGPIPAKWRGGSSK-----KVPCNRKLIGA 180
+ G I+ +DTGVWPES+SF D G GP+P+ W+G CN+KLIGA
Sbjct: 506 FPASGAGGEVIVGVLDTGVWPESKSFDDTGFGPVPSTWKGECESGTNFTAANCNKKLIGA 685
Query: 181 RFFSDAYERYNGKLPTS--QRTARDFVGHGTHTLSTAGGNFVPGASIFNIGNGTIKGGSP 238
R+FS E G + + ++ RD GHGTHT STA G+ V GAS+F GT +G +P
Sbjct: 686 RYFSKGAEAMLGPIDETAESKSPRDDDGHGTHTASTAAGSVVSGASLFGYAAGTARGMAP 865
Query: 239 RARVATYKVCWSLTDAASCFGADVLSAIDQAIDDGVDIISVSAGGPSSTNSEEIFTDEVS 298
ARVA YKVCW CF +D+L+AID+AI D V+++S+S GG + + + D V+
Sbjct: 866 HARVAAYKVCWK----GGCFSSDILAAIDKAIADNVNVLSLSLGGGMA----DYYRDSVA 1021
Query: 299 IGAFHALARNILLVASAGNEGPTPGSVVNVAPWVFTVAASTIDRDFSSTITIGD 352
IGAF A+ + IL+ SAGN GP+ S+ NVAPW+ TV A T+DRDF + + +G+
Sbjct: 1022IGAFAAMEKGILVSCSAGNAGPSAYSLSNVAPWITTVGAGTLDRDFPALVELGN 1183
>CN824981
Length = 726
Score = 216 bits (550), Expect = 8e-57
Identities = 115/235 (48%), Positives = 153/235 (64%), Gaps = 8/235 (3%)
Frame = +1
Query: 143 GVWPESRSFSDRGIGPIPAKWRG-----GSSKKVPCNRKLIGARFFSDAYERYNGKLP-- 195
GVWPE +S D G+ P+P+ W+G + CNRKLIGARFFS YE G +
Sbjct: 4 GVWPELKSLDDTGLSPVPSTWKGQCEAGNNMNSSSCNRKLIGARFFSKGYEATLGPIDVS 183
Query: 196 TSQRTARDFVGHGTHTLSTAGGNFVPGASIFNIGNGTIKGGSPRARVATYKVCWSLTDAA 255
T R+ARD GHG+HTL+TA G+ V GAS+F + +GT +G + +ARVA YKVCW
Sbjct: 184 TESRSARDDDGHGSHTLTTAAGSAVAGASLFGLASGTARGMATQARVAAYKVCW----LG 351
Query: 256 SCFGADVLSAIDQAIDDGVDIISVSAGGPSSTNSEEIFTDEVSIGAFHALARNILLVASA 315
CF +D+ + ID+AI+DGV+IIS+S GG S+ + F D ++IGAF A + IL+ SA
Sbjct: 352 GCFSSDIAAGIDKAIEDGVNIISMSIGGSSA----DYFRDIIAIGAFTANSHGILVSTSA 519
Query: 316 GNEGPTPGSVVNVAPWVFTVAASTIDRDFSSTITIGDQIIR-GASLFVDLPPNQS 369
GN GP+P S+ N APW+ TV A TIDRDF + IT+G+ I GASL+ P + S
Sbjct: 520 GNGGPSPSSLSNTAPWITTVGAGTIDRDFPAYITLGNNITHTGASLYRGKPLSDS 684
>TC13087 similar to UP|Q9FIF8 (Q9FIF8) Serine protease-like protein, partial
(31%)
Length = 1036
Score = 209 bits (531), Expect = 1e-54
Identities = 143/373 (38%), Positives = 205/373 (54%), Gaps = 9/373 (2%)
Frame = +2
Query: 200 TARDFVGHGTHTLSTAGGNFVPGASIFNIGNGTIKGGSPRARVATYKVCWSLTDAASCFG 259
T RD GHG+HT STA GN V GAS + + GT +G P AR+A YKVC+ C
Sbjct: 20 TVRDIEGHGSHTASTAAGNTVIGASFYGLAQGTARGAVPSARIAAYKVCYP---NHGCDD 190
Query: 260 ADVLSAIDQAIDDGVDIISVSAGGPSSTNSEEIFTDEVSIGAFHALARNILLVASAGNEG 319
A++L+A D AI DGV+I+SVS G + ++ +D V+IG+FHA+ R IL + +AGN G
Sbjct: 191 ANILAAFDDAIADGVNILSVSLG---TNYCVDLLSDTVAIGSFHAMERGILTLQAAGNSG 361
Query: 320 PTPGSVVNVAPWVFTVAASTIDRDFSSTITIGD-QIIRGASLFVDLPPNQSFTLVNSIDA 378
P S+ +VAPW+ TVAASTIDR F + +G+ + G S+ + F + A
Sbjct: 362 PASPSICSVAPWLLTVAASTIDRQFIDKVLLGNGMTLIGRSVNAVVSNGTKFPI-----A 526
Query: 379 KFSNATTRDARFC-RPRTLDPSKVKGKIVACAREGKIKSVAEGQEALSAGAKGMFLENQP 437
K++ + R F + L VKGK+V C EG++ A A GA G E+
Sbjct: 527 KWNASDERCKGFSDLCQCLGSESVKGKVVLC--EGRMHDEA----AYLDGAVGSITED-- 682
Query: 438 KVSGNTLLSEPHVLSTVGGNGQAAITAPPRLGVTATD--TIESGTKIRFSQAITLIGR-- 493
L+ P N + +T P + + D +++ T + ++
Sbjct: 683 -------LTNP-------DNDVSFVTYFPSVNLNPKDYAIVQNYTNSAKNPKAEILKSEI 820
Query: 494 ---KPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASASNLLTDNRRGFPFN 550
+ AP + SFSSRGPN + P I+KPDV+APGV+ILAA+S A+ S +D R+ +N
Sbjct: 821 FKDRSAPKVVSFSSRGPNPLIPEIMKPDVSAPGVDILAAWSPEAAPSEYSSDKRK-VRYN 997
Query: 551 VMQGTSMSCPHVA 563
V+ GTSM+CPHVA
Sbjct: 998 VVSGTSMACPHVA 1036
>BP066980
Length = 472
Score = 174 bits (441), Expect = 4e-44
Identities = 82/103 (79%), Positives = 94/103 (90%)
Frame = +3
Query: 21 QCYIVYMGAHSHGPTPTSVDLETATSSHYDLLGSIVGSKEEAKEAIIYSYNKQINGFAAM 80
+CYIVY+GAHSHGPTP+SVDLETATSSHYDLLGS++GS E+AKEAI+YSYNK INGFAA+
Sbjct: 162 KCYIVYLGAHSHGPTPSSVDLETATSSHYDLLGSVLGSHEKAKEAILYSYNKHINGFAAL 341
Query: 81 LEEEEAAQLAKNPKVVSVFLSKEHKLHTTRSWEFLGLHGNDIN 123
LEEEEAA++ KNPKVVSVF+SKEH + T RSWEFLGL GN N
Sbjct: 342 LEEEEAARIGKNPKVVSVFVSKEH*IDTNRSWEFLGLRGNGGN 470
>TC13765 similar to UP|Q9LLL8 (Q9LLL8) Subtilisin-type serine endopeptidase
XSP1, partial (20%)
Length = 635
Score = 131 bits (330), Expect = 3e-31
Identities = 83/229 (36%), Positives = 120/229 (52%), Gaps = 5/229 (2%)
Frame = +1
Query: 365 PPNQSFTLVNSIDAKFSNATTRDARFCRPRTLDPSKVKGKIVAC-----AREGKIKSVAE 419
P + ++L+N IDA + + DA FC +L P+KVKGK+V C EG +K
Sbjct: 1 PKRKEYSLINGIDAAKDSKSKDDAGFCYEDSLQPNKVKGKLVYCKLGNWGTEGVVKKF-- 174
Query: 420 GQEALSAGAKGMFLENQPKVSGNTLLSEPHVLSTVGGNGQAAITAPPRLGVTATDTIESG 479
G G +E+ + P A +G + T+ I+S
Sbjct: 175 -------GGIGSIMESDQYPDLAQIFMAP------------ATILNHTIGESVTNYIKST 297
Query: 480 TKIRFSQAITLIGRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASASNL 539
T + PAP +A+FSSRGPN +LKPD+ APG++ILA+Y+L S +
Sbjct: 298 RSPSAVIYKTHEEKCPAPFVATFSSRGPNPGSHNVLKPDIAAPGIDILASYTLRKSITGS 477
Query: 540 LTDNRRGFPFNVMQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTT 588
D + F+++ GTSM+CPHVAG A +K+ HPNW+PAAI+SAI+TT
Sbjct: 478 EGDTQFS-EFSLLSGTSMACPHVAGVAAYVKSFHPNWTPAAIRSAIITT 621
>AU088875
Length = 322
Score = 119 bits (299), Expect = 1e-27
Identities = 61/77 (79%), Positives = 71/77 (91%), Gaps = 1/77 (1%)
Frame = +3
Query: 290 EEIFTDEVSIGAFHALARNILLVASAGNEGPTPGSVVNVAPWVFTVAASTIDRDFSSTIT 349
EEIFTDEVSIG+FHALA+NIL+VASAGN+GP G+V+NVAPWVFTVAASTID +FSSTIT
Sbjct: 3 EEIFTDEVSIGSFHALAKNILVVASAGNDGPALGTVLNVAPWVFTVAASTIDXEFSSTIT 182
Query: 350 IG-DQIIRGASLFVDLP 365
+G +Q I GASLFV+LP
Sbjct: 183 LGNNQQITGASLFVNLP 233
>AV421923
Length = 463
Score = 119 bits (298), Expect = 1e-27
Identities = 63/112 (56%), Positives = 83/112 (73%), Gaps = 3/112 (2%)
Frame = +2
Query: 487 AITLIGRK---PAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASASNLLTDN 543
+I+ IG + PAPV+A+FSSRGP+ V ++KPDVTAPGV+ILAA+ S S L D
Sbjct: 17 SISFIGTRFGDPAPVVAAFSSRGPSIVGLDVIKPDVTAPGVHILAAWPSKTSPSMLKNDK 196
Query: 544 RRGFPFNVMQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTVITYLSK 595
RR FN++ GTSMSCPHV+G A LI+++H +WSPAA+KSA+MTT T +K
Sbjct: 197 RRVL-FNIISGTSMSCPHVSGVAALIRSVHRDWSPAAVKSALMTTAYTLNNK 349
>AV420480
Length = 412
Score = 117 bits (293), Expect = 5e-27
Identities = 60/100 (60%), Positives = 73/100 (73%)
Frame = +3
Query: 489 TLIGRKPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASASNLLTDNRRGFP 548
T+IG AP +A+FS+RGP+ P ILKPDV APGVNI+AA+ ++L D RR
Sbjct: 9 TVIGNSRAPAVATFSARGPSFTNPSILKPDVVAPGVNIIAAWPQNLGPTSLPQDLRR-VN 185
Query: 549 FNVMQGTSMSCPHVAGTAGLIKTLHPNWSPAAIKSAIMTT 588
F+VM GTSMSCPHV+G A L+ + HP WSPAAIKSAIMTT
Sbjct: 186 FSVMSGTSMSCPHVSGIAALVHSAHPKWSPAAIKSAIMTT 305
>AU088794
Length = 433
Score = 112 bits (280), Expect = 2e-25
Identities = 57/98 (58%), Positives = 70/98 (71%)
Frame = +2
Query: 498 VMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASASNLLTDNRRGFPFNVMQGTSM 557
V+ASFS+RGPN P ILKPDV PG+NILAA+ S + +D RR FN++ GTSM
Sbjct: 2 VVASFSARGPNPESPEILKPDVIXPGLNILAAWPDXXGPSGVPSDVRRT-EFNILSGTSM 178
Query: 558 SCPHVAGTAGLIKTLHPNWSPAAIKSAIMTTVITYLSK 595
+CPHV+G A L+K HP WSPAAIKSA+MTT T +K
Sbjct: 179 ACPHVSGLAALLKAAHPXWSPAAIKSALMTTAYTVDNK 292
>TC17430 weakly similar to UP|Q9ZTT3 (Q9ZTT3) Subtilisin-like protease C1,
partial (12%)
Length = 729
Score = 102 bits (255), Expect = 1e-22
Identities = 60/153 (39%), Positives = 87/153 (56%)
Frame = +1
Query: 23 YIVYMGAHSHGPTPTSVDLETATSSHYDLLGSIVGSKEEAKEAIIYSYNKQINGFAAMLE 82
YIVYMG H G P S + S H + ++GS E + +I++SY K NGF L
Sbjct: 73 YIVYMGDHPKGMDPAS-----SPSLHMAMAQKVIGSNSEPR-SILHSY-KSFNGFVMKLT 231
Query: 83 EEEAAQLAKNPKVVSVFLSKEHKLHTTRSWEFLGLHGNDINSAWQKGRFGENTIIANIDT 142
EEE ++A+ +VVSV ++ LHTT+SW+F+G ++ + + I+ +DT
Sbjct: 232 EEEKERMAEMDEVVSVIPDSKYILHTTKSWDFIGFPQQVTRASME-----SDIIVGVVDT 396
Query: 143 GVWPESRSFSDRGIGPIPAKWRGGSSKKVPCNR 175
G+ PES+SFSD G GP P K + GSS CN+
Sbjct: 397 GIGPESKSFSDEGFGPPPKKGK-GSSHNFTCNK 492
>BP033910
Length = 538
Score = 100 bits (249), Expect = 7e-22
Identities = 54/138 (39%), Positives = 79/138 (57%)
Frame = +1
Query: 23 YIVYMGAHSHGPTPTSVDLETATSSHYDLLGSIVGSKEEAKEAIIYSYNKQINGFAAMLE 82
YIVYMG P + L +H++LL + +G+K+ A++ ++SY K NGF A L
Sbjct: 148 YIVYMGE-----LPAARTLHAMQENHHNLLTTAIGNKQLARQFKMHSYGKSFNGFVARLL 312
Query: 83 EEEAAQLAKNPKVVSVFLSKEHKLHTTRSWEFLGLHGNDINSAWQKGRFGENTIIANIDT 142
EA +L ++ VVSVF + KLHTTRSW+FLG+ N + N ++ +DT
Sbjct: 313 PNEAEKLLEDDNVVSVFPNTRRKLHTTRSWDFLGMPLN----VRRNPAMESNMVVGVLDT 480
Query: 143 GVWPESRSFSDRGIGPIP 160
G+W + SF+D G GP P
Sbjct: 481 GIWVDCPSFNDTGYGPPP 534
>TC19089 similar to UP|Q9ZTT3 (Q9ZTT3) Subtilisin-like protease C1, partial
(16%)
Length = 600
Score = 100 bits (248), Expect = 9e-22
Identities = 46/83 (55%), Positives = 58/83 (69%)
Frame = +1
Query: 506 GPNQVQPYILKPDVTAPGVNILAAYSLFASASNLLTDNRRGFPFNVMQGTSMSCPHVAGT 565
GPN + P ILKPD+ APGV +LAA+S S + D R+ P+N + GTSM+CPH A
Sbjct: 16 GPNPITPNILKPDIAAPGVTVLAAWSPLNPISEVEGDKRK-LPYNAISGTSMACPHAAAA 192
Query: 566 AGLIKTLHPNWSPAAIKSAIMTT 588
A +K+ HPNWSPA IKSA+MTT
Sbjct: 193 AAYVKSFHPNWSPAMIKSALMTT 261
>AV408408
Length = 429
Score = 94.7 bits (234), Expect = 4e-20
Identities = 62/149 (41%), Positives = 80/149 (53%)
Frame = +3
Query: 383 ATTRDARFCRPRTLDPSKVKGKIVACAREGKIKSVAEGQEALSAGAKGMFLENQPKVSGN 442
+ T + C TL P KV GKIV C R G V +GQ AG GM L N +G
Sbjct: 6 SNTTNGNLCMMGTLSPDKVAGKIVMCDR-GMNARVQKGQVVKGAGGLGMVLSNTA-ANGE 179
Query: 443 TLLSEPHVLSTVGGNGQAAITAPPRLGVTATDTIESGTKIRFSQAITLIGRKPAPVMASF 502
L+++ H+L T +A L T+ KI F T +G +P+PV+A+F
Sbjct: 180 ELVADAHLLPTSAVGEKAGNAIKKYLFSDPKATV----KILFEG--TKVGIQPSPVVAAF 341
Query: 503 SSRGPNQVQPYILKPDVTAPGVNILAAYS 531
SSRGPN + P ILKPD+ APGVNILA +S
Sbjct: 342 SSRGPNSITPQILKPDLIAPGVNILAGWS 428
>TC19209 similar to GB|BAB11244.1|10177874|AB010074 serine protease-like
protein {Arabidopsis thaliana;} , partial (12%)
Length = 553
Score = 91.7 bits (226), Expect = 3e-19
Identities = 53/148 (35%), Positives = 78/148 (51%), Gaps = 4/148 (2%)
Frame = +1
Query: 18 FLFQCYIVYMGAHSHGPTPTSVDLETATSSHYDLLGSIVGSK----EEAKEAIIYSYNKQ 73
FL + YI+ + + S LE +S +L + S+ EE +E IIYSY+
Sbjct: 106 FLKKTYIIQIDNSAMPAGTFSTHLEWYSSKVKSVLSKSLESETISSEEEEERIIYSYHTA 285
Query: 74 INGFAAMLEEEEAAQLAKNPKVVSVFLSKEHKLHTTRSWEFLGLHGNDINSAWQKGRFGE 133
+G AA L EEA +L VV++F ++LHTTRS FLGL W +
Sbjct: 286 FHGVAAKLTPEEAQKLESQEGVVAIFPETRYQLHTTRSPTFLGLEKMQSTEIWSEKLSNH 465
Query: 134 NTIIANIDTGVWPESRSFSDRGIGPIPA 161
+ ++ DTG+WPES+SF+D G P+P+
Sbjct: 466 DVVVGVSDTGIWPESQSFNDTGFKPVPS 549
>AV426475
Length = 300
Score = 88.2 bits (217), Expect = 3e-18
Identities = 43/77 (55%), Positives = 56/77 (71%)
Frame = +2
Query: 494 KPAPVMASFSSRGPNQVQPYILKPDVTAPGVNILAAYSLFASASNLLTDNRRGFPFNVMQ 553
KP+PV+A+FSSRGPN + P ILKPD+ APGVNILA ++ + L D R FN++
Sbjct: 71 KPSPVVAAFSSRGPNGLTPKILKPDLIAPGVNILAGWTGAIGPTGLPVDTRH-VSFNIIS 247
Query: 554 GTSMSCPHVAGTAGLIK 570
GTSMSCPHV+G A ++K
Sbjct: 248 GTSMSCPHVSGLAAILK 298
>AV418734
Length = 287
Score = 82.0 bits (201), Expect = 2e-16
Identities = 44/78 (56%), Positives = 56/78 (71%)
Frame = +1
Query: 519 VTAPGVNILAAYSLFASASNLLTDNRRGFPFNVMQGTSMSCPHVAGTAGLIKTLHPNWSP 578
VTAPG+NILAA+S +A N+ FN++ GTSM+CPHV G A L+K +HP+WSP
Sbjct: 4 VTAPGLNILAAWS--PAAGNM---------FNIVSGTSMACPHVTGIATLVKAVHPSWSP 150
Query: 579 AAIKSAIMTTVITYLSKY 596
+AIKSAIMTT T L K+
Sbjct: 151 SAIKSAIMTTA-TILDKH 201
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.134 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,007,697
Number of Sequences: 28460
Number of extensions: 134722
Number of successful extensions: 952
Number of sequences better than 10.0: 64
Number of HSP's better than 10.0 without gapping: 874
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 896
length of query: 609
length of database: 4,897,600
effective HSP length: 96
effective length of query: 513
effective length of database: 2,165,440
effective search space: 1110870720
effective search space used: 1110870720
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Medicago: description of AC133779.10


