

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133571.8 - phase: 0
(495 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
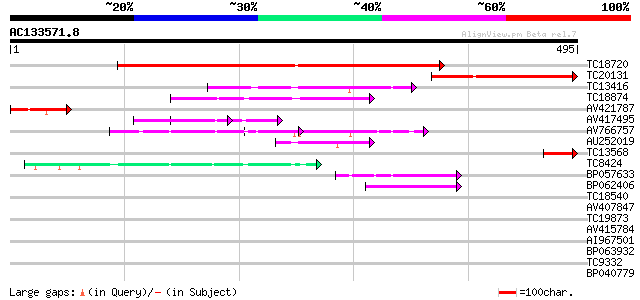
Score E
Sequences producing significant alignments: (bits) Value
TC18720 weakly similar to UP|SCWA_YEAST (Q04951) Probable family... 390 e-109
TC20131 174 3e-44
TC13416 weakly similar to GB|AAP21292.1|30102748|BT006484 At1g67... 83 1e-16
TC18874 similar to UP|Q9FJJ0 (Q9FJJ0) Genomic DNA, chromosome 5,... 62 2e-10
AV421787 61 5e-10
AV417495 59 2e-09
AV766757 58 3e-09
AU252019 49 1e-06
TC13568 47 7e-06
TC8424 weakly similar to GB|BAC22265.1|24414015|AP003803 arm rep... 45 4e-05
BP057633 42 3e-04
BP062406 41 4e-04
TC18540 similar to UP|Q944I3 (Q944I3) AT3g01400/T13O15_4, partia... 39 0.003
AV407847 38 0.004
TC19873 weakly similar to GB|AAP12859.1|30017251|BT006210 At3g16... 37 0.010
AV415784 34 0.062
AI967501 32 0.24
BP063932 30 0.69
TC9332 similar to UP|Q9ARG5 (Q9ARG5) Plasma membrane H+ ATPase, ... 29 2.0
BP040779 29 2.0
>TC18720 weakly similar to UP|SCWA_YEAST (Q04951) Probable family 17
glucosidase SCW10 precursor (Soluble cell wall protein
10) , partial (7%)
Length = 871
Score = 390 bits (1002), Expect = e-109
Identities = 214/286 (74%), Positives = 239/286 (82%), Gaps = 1/286 (0%)
Frame = +3
Query: 95 ETNAESKKKEETLTEMKHVVKDL-RGEDSTKRRIAAARVRSLTKEDSEARGSLAMLGAIS 153
E AE+K KE+ L EMK +V +L +G+D+ KRR AAA VR L KEDS RG+LAMLGAI
Sbjct: 18 EAEAETKNKEDALEEMKRLVAELHQGDDTVKRREAAATVRMLAKEDSHVRGTLAMLGAIP 197
Query: 154 PLVGMLDSEDLHSQIDSLYALLNLGIANDANKAAIVKIGAVHKMLKLIESPCVVDSSVSE 213
PL MLDSEDL SQ SLYALLNLGI NDANKAAIVK+G+VHKMLKLIESP +DS VSE
Sbjct: 198 PLGAMLDSEDLDSQKASLYALLNLGIGNDANKAAIVKVGSVHKMLKLIESPEGLDSRVSE 377
Query: 214 AIVANFLGLSALDSNKPIIGSSGAIPFLVRILKNLDNSSKSSSQVKQDALRALYNLSINQ 273
AIVANFLGLSALDSNKPIIGSS A+PFLVR L++ D K+SSQ KQDALRALYNLSI
Sbjct: 378 AIVANFLGLSALDSNKPIIGSSAAVPFLVRTLQSSD--VKTSSQAKQDALRALYNLSIFP 551
Query: 274 TNISFVLETDLVVFLINSIEDMEVSERVLSILSNLVSSPEGRKAISAVKDAITVLVDVLN 333
NISF+LET LV FLINSI DMEV+ER LSILSNLVS+ EGRKAISAV D +LVDVLN
Sbjct: 552 GNISFILETHLVPFLINSIGDMEVTERSLSILSNLVSTREGRKAISAVPDTFPILVDVLN 731
Query: 334 WTDSPECQEKASYILMIMAHKAYADRQAMIEAGIVSSLLELTLVGT 379
WTDSPECQEK SY+LM+MAHK++ DRQ MIEAGIVSSLLEL+L+GT
Sbjct: 732 WTDSPECQEKTSYVLMVMAHKSFGDRQGMIEAGIVSSLLELSLLGT 869
>TC20131
Length = 550
Score = 174 bits (441), Expect = 3e-44
Identities = 98/128 (76%), Positives = 105/128 (81%), Gaps = 1/128 (0%)
Frame = +1
Query: 369 SSLLELTLVGTALAQKRASRILQCFRLDKGKQVSRSCDGGNLGLTVSAPICASSSSLVKT 428
SSLLEL+L+GT LAQKRASR+L R+DKGK VS SC GGNLG TVSAPIC SSSS K
Sbjct: 1 SSLLELSLLGTTLAQKRASRMLGSLRVDKGKYVSGSC-GGNLGATVSAPICGSSSSCAKP 177
Query: 429 DGGGKECLMEEVNMM-SDEKKAVKQLVQQSLQNNMMKIVKRANLRQDFVPSERFASLTSS 487
DGGGKE E+ +MM SDEKKAVKQLVQQSLQNNM KIVKRANL D VPS+ F SLTSS
Sbjct: 178 DGGGKEGSEEDEDMMMSDEKKAVKQLVQQSLQNNMRKIVKRANLPHDIVPSDHFRSLTSS 357
Query: 488 STSKSLPF 495
STSKSLPF
Sbjct: 358 STSKSLPF 381
>TC13416 weakly similar to GB|AAP21292.1|30102748|BT006484 At1g67530
{Arabidopsis thaliana;}, partial (23%)
Length = 557
Score = 82.8 bits (203), Expect = 1e-16
Identities = 58/186 (31%), Positives = 103/186 (55%), Gaps = 3/186 (1%)
Frame = +2
Query: 173 ALLNLGIANDANKAAIVKIGAVHKMLKLIESPCVVDSSVSEAIVANFLGLSALDSNKPII 232
AL NL + N+ NK ++ G + + ++I + S+ A +L LS L+ KP+I
Sbjct: 29 ALFNLAVNNNGNKEMMLSAGVLPVLEEMISNTSSYGSAT-----ALYLNLSCLEEAKPMI 193
Query: 233 GSSGAIPFLVRILKNLDNSSKSSSQVKQDALRALYNLSINQTNISFVLETDLVVFLINSI 292
G+S A+ FL ++L+ S S Q KQD+L ALYNLS TNI ++L + ++ L + +
Sbjct: 194 GTSQAVQFLTQLLQ-----SDSDIQCKQDSLHALYNLSTVSTNIPYLL*SGIINGLQSLL 358
Query: 293 EDM---EVSERVLSILSNLVSSPEGRKAISAVKDAITVLVDVLNWTDSPECQEKASYILM 349
D +E+ +++L NL +S GR+ + + I+ L +L+ QE+A+ L+
Sbjct: 359 VDQGDCVWTEKCIAVLINLATSQAGREEMVSAPGLISALASILD-IGELIVQEQAASCLL 535
Query: 350 IMAHKA 355
I+ +++
Sbjct: 536 ILCNRS 553
>TC18874 similar to UP|Q9FJJ0 (Q9FJJ0) Genomic DNA, chromosome 5, TAC
clone:K19B1, partial (33%)
Length = 543
Score = 62.4 bits (150), Expect = 2e-10
Identities = 49/178 (27%), Positives = 93/178 (51%)
Frame = +2
Query: 141 EARGSLAMLGAISPLVGMLDSEDLHSQIDSLYALLNLGIANDANKAAIVKIGAVHKMLKL 200
EAR SL +S L +++S + Q++++ +L+NL + NK IV+ G V ++ +
Sbjct: 14 EARVSLCTPRLLSALRLLVESRYVVVQVNAVASLVNLSLEK-TNKVRIVRSGFVPFLIDV 190
Query: 201 IESPCVVDSSVSEAIVANFLGLSALDSNKPIIGSSGAIPFLVRILKNLDNSSKSSSQVKQ 260
++ S E L+ D NK IG GA+ L+ L++ S + +
Sbjct: 191 LKGGF---SESQEHAAGALFSLALDDDNKMAIGVLGALQPLMHALRS------ESERTRH 343
Query: 261 DALRALYNLSINQTNISFVLETDLVVFLINSIEDMEVSERVLSILSNLVSSPEGRKAI 318
D+ ALY+L++ Q+N +++ +V L++ + + ++ RVL +L N+ + EGR A+
Sbjct: 344 DSALALYHLTLVQSNRVKLVKLGVVPTLLSMVANGNLASRVLLVLCNVAACVEGRTAM 517
>AV421787
Length = 250
Score = 60.8 bits (146), Expect = 5e-10
Identities = 33/61 (54%), Positives = 41/61 (67%), Gaps = 7/61 (11%)
Frame = +1
Query: 1 MAKCHRSNVDSIVLHHRHHAKNTTAGNFRF------SASSFRRIIFDALSCGGASR-HHR 53
MA CHR+NV +IVL H H K TT + F SA+SFRR++ DA+SCG +SR HHR
Sbjct: 49 MANCHRNNVGNIVLGH-HPTKTTTTFSTHFRLWNSLSAASFRRVVLDAVSCGASSRYHHR 225
Query: 54 H 54
H
Sbjct: 226 H 228
>AV417495
Length = 316
Score = 58.5 bits (140), Expect = 2e-09
Identities = 36/86 (41%), Positives = 52/86 (59%)
Frame = +3
Query: 109 EMKHVVKDLRGEDSTKRRIAAARVRSLTKEDSEARGSLAMLGAISPLVGMLDSEDLHSQI 168
+++ +V+ LR D +R A +R L K + + R ++A GAIS LV +L S D Q
Sbjct: 57 QVRGLVEGLRSSDLDTQREATVEIRFLAKHNMDNRIAIANCGAISLLVELLKSTDTRIQE 236
Query: 169 DSLYALLNLGIANDANKAAIVKIGAV 194
+++ ALLNL I ND NKAAI GA+
Sbjct: 237 NAVTALLNLSI-NDNNKAAIANAGAI 311
Score = 44.7 bits (104), Expect = 4e-05
Identities = 33/99 (33%), Positives = 52/99 (52%), Gaps = 1/99 (1%)
Frame = +3
Query: 141 EARGSLAMLGA-ISPLVGMLDSEDLHSQIDSLYALLNLGIANDANKAAIVKIGAVHKMLK 199
E R L+ + A + LV L S DL +Q ++ + L N N+ AI GA+ +++
Sbjct: 24 ETRADLSGIEAQVRGLVEGLRSSDLDTQREATVEIRFLAKHNMDNRIAIANCGAISLLVE 203
Query: 200 LIESPCVVDSSVSEAIVANFLGLSALDSNKPIIGSSGAI 238
L++S D+ + E V L LS D+NK I ++GAI
Sbjct: 204 LLKS---TDTRIQENAVTALLNLSINDNNKAAIANAGAI 311
>AV766757
Length = 601
Score = 58.2 bits (139), Expect = 3e-09
Identities = 53/169 (31%), Positives = 79/169 (46%)
Frame = +2
Query: 88 LLNIQVHETNAESKKKEETLTEMKHVVKDLRGEDSTKRRIAAARVRSLTKEDSEARGSLA 147
LLN+ ++E N + + HV LR R AAA + SL+ D E + ++
Sbjct: 56 LLNLSIYENNKGCIVSSGAVPGIVHV---LRKGSMEARENAAATLFSLSVVD-ENKVTIG 223
Query: 148 MLGAISPLVGMLDSEDLHSQIDSLYALLNLGIANDANKAAIVKIGAVHKMLKLIESPCVV 207
GAI PLV +L + D+ AL NL I NK V+ G + ++KL+ P
Sbjct: 224 SSGAIPPLVTLLSEGTQRGKKDAATALFNLCI-YQGNKGKAVRAGVIPTLMKLLTEPS-- 394
Query: 208 DSSVSEAIVANFLGLSALDSNKPIIGSSGAIPFLVRILKNLDNSSKSSS 256
V EA+ A LS+ K IG++ A+P LV + N +K +S
Sbjct: 395 GGMVDEAL-AILAILSSHPDGKAAIGAADAVPILVEFIGNGSPRNKENS 538
Score = 57.8 bits (138), Expect = 4e-09
Identities = 53/197 (26%), Positives = 91/197 (45%), Gaps = 37/197 (18%)
Frame = +2
Query: 206 VVDSSVSEAIVANFLGLSALDSNKPIIGSSGAIPFLVRILKN------------------ 247
V DS E V L LS ++NK I SSGA+P +V +L+
Sbjct: 17 VPDSRTQEHAVTALLNLSIYENNKGCIVSSGAVPGIVHVLRKGSMEARENAAATLFSLSV 196
Query: 248 LDNS-----------------SKSSSQVKQDALRALYNLSINQTNISFVLETDLVVFLIN 290
+D + S+ + + K+DA AL+NL I Q N + ++ L+
Sbjct: 197 VDENKVTIGSSGAIPPLVTLLSEGTQRGKKDAATALFNLCIYQGNKGKAVRAGVIPTLMK 376
Query: 291 SIEDME--VSERVLSILSNLVSSPEGRKAISAVKDAITVLVDVLNWTDSPECQEKASYIL 348
+ + + + L+IL+ L S P+G+ AI A DA+ +LV+ + SP +E ++ +L
Sbjct: 377 LLTEPSGGMVDEALAILAILSSHPDGKAAIGAA-DAVPILVEFIG-NGSPRNKENSAAVL 550
Query: 349 MIMAHKAYADRQAMIEA 365
+ H + D+Q + +A
Sbjct: 551 V---HLSSGDQQYLAQA 592
>AU252019
Length = 329
Score = 49.3 bits (116), Expect = 1e-06
Identities = 32/89 (35%), Positives = 52/89 (57%), Gaps = 3/89 (3%)
Frame = +2
Query: 233 GSSGAIPFLVRILKNLDNSSKSSSQVKQDALRALYNLSINQTNISFVLETDL---VVFLI 289
G+S A+ FL+ IL+ S + Q K D+L ALYNLS +NIS++L + + + L+
Sbjct: 56 GTSQAVQFLIHILQ-----SNTEVQCKLDSLHALYNLSTVSSNISYLLSSGIMNGLQSLL 220
Query: 290 NSIEDMEVSERVLSILSNLVSSPEGRKAI 318
+D +E+ +++L NL S GR+ I
Sbjct: 221 VGQDDSMWTEKCIAVLINLAVSQVGREEI 307
>TC13568
Length = 426
Score = 47.0 bits (110), Expect = 7e-06
Identities = 23/29 (79%), Positives = 24/29 (82%)
Frame = +3
Query: 467 KRANLRQDFVPSERFASLTSSSTSKSLPF 495
KRANL D VPS+ F SLTSSSTSKSLPF
Sbjct: 3 KRANLPHDIVPSDHFRSLTSSSTSKSLPF 89
>TC8424 weakly similar to GB|BAC22265.1|24414015|AP003803 arm repeat
containing protein homolog-like {Oryza sativa (japonica
cultivar-group);} , partial (49%)
Length = 1586
Score = 44.7 bits (104), Expect = 4e-05
Identities = 67/290 (23%), Positives = 110/290 (37%), Gaps = 31/290 (10%)
Frame = +3
Query: 14 LHHRHHAK---------------------NTTAGNFRFSASSFRRIIFDA---LSCGGAS 49
LHH HH+ N++ +F ++ F + D+ L G +
Sbjct: 123 LHHHHHSSKTQRHIGRSMRTIRSNFFQDDNSSTCSFTEKSTCFSENLTDSVVDLRLGELA 302
Query: 50 RHHRHYHREE-------EISSVASTATVKELRGEVQEEKTEKLLDLLNIQVHETNAESKK 102
HR ++S S + QE LL+L N E+KK
Sbjct: 303 TRSNKSHRSSFADEDLLDLSQAFSDFSACSSDPWTQEHAVTALLNL------SLNDENKK 464
Query: 103 KEETLTEMKHVVKDLRGEDSTKRRIAAARVRSLTKEDSEARGSLAMLGAISPLVGMLDSE 162
+K ++ L+ T ++ AA + SL + E R S+ GAI PLV +L +
Sbjct: 465 LITNAGAVKSLIYVLKTGTETSKQNAACALLSLALVE-ENRSSIGASGAIPPLVSLLING 641
Query: 163 DLHSQIDSLYALLNLGIANDANKAAIVKIGAVHKMLKLIESPCVVDSSVSEAIVANFLGL 222
+ D+L L L + NK V G V ++ L+ + ++E + L
Sbjct: 642 SSRGKKDALTTLYKL-CSVKLNKERAVNAGVVKPLVDLVAEQ---GTGLAEKAMVVLNSL 809
Query: 223 SALDSNKPIIGSSGAIPFLVRILKNLDNSSKSSSQVKQDALRALYNLSIN 272
+A+ K I G I LV ++ D S K K+ A+ L L ++
Sbjct: 810 AAVQDGKDAIVEEGGIAALVEAIE--DGSVKG----KEFAVLTLLQLCVD 941
>BP057633
Length = 570
Score = 41.6 bits (96), Expect = 3e-04
Identities = 32/110 (29%), Positives = 63/110 (57%)
Frame = -2
Query: 285 VVFLINSIEDMEVSERVLSILSNLVSSPEGRKAISAVKDAITVLVDVLNWTDSPECQEKA 344
+V L+N+ + M ++ +++LSNL + EGR I A + I +LV+++ + S +E A
Sbjct: 548 LVQLMNTADGMV--DKAVALLSNLSTITEGRSEI-AREGGIPLLVEIIE-SGSQRGKENA 381
Query: 345 SYILMIMAHKAYADRQAMIEAGIVSSLLELTLVGTALAQKRASRILQCFR 394
+ IL+ + + +++ G V L+ L+ GT A+++A ++L FR
Sbjct: 380 ASILLQLCLHSSKFCTLVLQEGAVPPLVALSQSGTPRAKEKAQQLLSHFR 231
>BP062406
Length = 510
Score = 41.2 bits (95), Expect = 4e-04
Identities = 27/84 (32%), Positives = 38/84 (45%)
Frame = -3
Query: 311 SPEGRKAISAVKDAITVLVDVLNWTDSPECQEKASYILMIMAHKAYADRQAMIEAGIVSS 370
S EGR AIS V I LV+ + + +L + R+ ++ G +
Sbjct: 508 SEEGRTAISLVDGGILTLVETVEDGSLVSTEHAVGALLSLCRSSRDKYRELILNEGAIPG 329
Query: 371 LLELTLVGTALAQKRASRILQCFR 394
LL LT+ GTA AQ RA +L R
Sbjct: 328 LLRLTVDGTAEAQNRARVLLDLLR 257
>TC18540 similar to UP|Q944I3 (Q944I3) AT3g01400/T13O15_4, partial (40%)
Length = 433
Score = 38.5 bits (88), Expect = 0.003
Identities = 42/144 (29%), Positives = 71/144 (49%), Gaps = 3/144 (2%)
Frame = +3
Query: 208 DSSVSEAIVANFLGLSALDSNKPIIGSSGAIPFLVRILKNLDNSSKSSSQVKQDALRALY 267
D + E V L LS D NK +I SSGAI LVR LK + K++A AL
Sbjct: 24 DLQLQEYGVTAILNLSLCDENKELIASSGAIKPLVRALK------AGTPTAKENAACALL 185
Query: 268 NLSINQTNISFVLETDLVVFLINSIE--DMEVSERVLSILSNLVSSPEGRKAISAVKDAI 325
LS + + + + + + L++ ++ + + + L +L S E + I AVK I
Sbjct: 186 RLSQAEESKAAIGRSGAIPLLVSLLQTGGIRGKKDASTALYSLCSVRENK--IRAVKAGI 359
Query: 326 -TVLVDVLNWTDSPECQEKASYIL 348
VLV+++ +S +K++Y++
Sbjct: 360 MKVLVELMADFES-NMVDKSAYVV 428
Score = 32.3 bits (72), Expect = 0.18
Identities = 33/123 (26%), Positives = 62/123 (49%)
Frame = +3
Query: 134 SLTKEDSEARGSLAMLGAISPLVGMLDSEDLHSQIDSLYALLNLGIANDANKAAIVKIGA 193
SL E+ E +A GAI PLV L + ++ ++ ALL L A + +KAAI + GA
Sbjct: 69 SLCDENKEL---IASSGAIKPLVRALKAGTPTAKENAACALLRLSQAEE-SKAAIGRSGA 236
Query: 194 VHKMLKLIESPCVVDSSVSEAIVANFLGLSALDSNKPIIGSSGAIPFLVRILKNLDNSSK 253
+ ++ L+++ + +A A + L ++ NK +G + LV ++ + +++
Sbjct: 237 IPLLVSLLQTGGI--RGKKDASTALY-SLCSVRENKIRAVKAGIMKVLVELMADFESNMV 407
Query: 254 SSS 256
S
Sbjct: 408 DKS 416
>AV407847
Length = 423
Score = 37.7 bits (86), Expect = 0.004
Identities = 31/85 (36%), Positives = 44/85 (51%), Gaps = 1/85 (1%)
Frame = +1
Query: 200 LIESPCVVDSSVSEAIVANFLGLSALDSN-KPIIGSSGAIPFLVRILKNLDNSSKSSSQV 258
L+ V D S A+V L+ DS+ + I +GAIP LVR L S S QV
Sbjct: 37 LVNKLNVSDLDESNAVVYELRVLAKTDSDSRACIAEAGAIPVLVRYLTT--GSHHPSLQV 210
Query: 259 KQDALRALYNLSINQTNISFVLETD 283
+A+ + NLSI + N + ++ETD
Sbjct: 211 --NAVTTILNLSILEANKTRIMETD 279
>TC19873 weakly similar to GB|AAP12859.1|30017251|BT006210 At3g16330
{Arabidopsis thaliana;}, partial (27%)
Length = 561
Score = 36.6 bits (83), Expect = 0.010
Identities = 22/83 (26%), Positives = 40/83 (47%), Gaps = 4/83 (4%)
Frame = +3
Query: 10 DSIVLHH--RHHAKNTTAGNFRFSASSFRRIIFDALSCGGASRHH--RHYHREEEISSVA 65
+++ LHH RH + + ++ FS S+ I+F + HH RH H E+IS+V
Sbjct: 252 NTLTLHHHRRHQHSSVSPQDYEFSCSNSPAIMFPIIKRHNKKYHHSRRHNHHYEDISTVH 431
Query: 66 STATVKELRGEVQEEKTEKLLDL 88
+ + E+ + + L+ L
Sbjct: 432 AVQKMLEMLNNSDKVEASPLVAL 500
>AV415784
Length = 242
Score = 33.9 bits (76), Expect = 0.062
Identities = 24/57 (42%), Positives = 32/57 (56%)
Frame = +1
Query: 133 RSLTKEDSEARGSLAMLGAISPLVGMLDSEDLHSQIDSLYALLNLGIANDANKAAIV 189
R L K E R +A GAI L +L S + +Q +S+ ALLNL I D NK+ I+
Sbjct: 4 RFLAKTGRENRAFIAEAGAIPYLRNLLSSHNAVAQENSVTALLNLSIF-DKNKSRIM 171
Score = 30.4 bits (67), Expect = 0.69
Identities = 17/56 (30%), Positives = 32/56 (56%)
Frame = +1
Query: 228 NKPIIGSSGAIPFLVRILKNLDNSSKSSSQVKQDALRALYNLSINQTNISFVLETD 283
N+ I +GAIP+L +L S ++ +++++ AL NLSI N S +++ +
Sbjct: 31 NRAFIAEAGAIPYLRNLL------SSHNAVAQENSVTALLNLSIFDKNKSRIMDEE 180
>AI967501
Length = 393
Score = 32.0 bits (71), Expect = 0.24
Identities = 19/71 (26%), Positives = 36/71 (49%)
Frame = +1
Query: 416 APICASSSSLVKTDGGGKECLMEEVNMMSDEKKAVKQLVQQSLQNNMMKIVKRANLRQDF 475
A +CA++S+L KTD G K +++ S+ K+ +Q+ Q+ Q+ + R ++
Sbjct: 52 AEVCATASTLSKTDSGSK----SDISKASERKQEQQQVPQKETQSKQICGRSRTKVQMQG 219
Query: 476 VPSERFASLTS 486
V R L +
Sbjct: 220 VAVGRAVDLNT 252
>BP063932
Length = 605
Score = 30.4 bits (67), Expect = 0.69
Identities = 27/105 (25%), Positives = 41/105 (38%), Gaps = 6/105 (5%)
Frame = -1
Query: 45 CGGASRHHRHYHREEEISSVASTATVKELRGEVQEEKTEKLLDLLNIQVHETNAESKKKE 104
C +RHH H R +EIS A E T + QVH T + +
Sbjct: 491 CKSLARHHSHGPRTQEISQCAQAHKDSRTWAHTME**TSR-------QVHRTGLXXQHSQ 333
Query: 105 ETLTEMKHVVKDLRGEDSTKRRI------AAARVRSLTKEDSEAR 143
TL + ++ +DS + A+ R SL+K+ S +R
Sbjct: 332 TTLIAL--AIRFRNSKDSVQTHSARHGSQASRRQSSLSKQRSSSR 204
>TC9332 similar to UP|Q9ARG5 (Q9ARG5) Plasma membrane H+ ATPase, partial
(25%)
Length = 960
Score = 28.9 bits (63), Expect = 2.0
Identities = 18/51 (35%), Positives = 24/51 (46%), Gaps = 3/51 (5%)
Frame = +1
Query: 49 SRHHRHYHREEEISSVASTATVKE---LRGEVQEEKTEKLLDLLNIQVHET 96
+ HH H E+ A A +KE L+G V+ K LD+ IQ H T
Sbjct: 586 NNHHEHSEIAEQAKRRAEAARLKELHTLKGHVESVVKLKGLDIETIQQHYT 738
>BP040779
Length = 496
Score = 28.9 bits (63), Expect = 2.0
Identities = 20/63 (31%), Positives = 35/63 (54%), Gaps = 1/63 (1%)
Frame = -1
Query: 365 AGIVSSLLELTLVGTAL-AQKRASRILQCFRLDKGKQVSRSCDGGNLGLTVSAPICASSS 423
+ +VS+L E L+ ++L + R + C R+ ++VS C G + + SA C+SS
Sbjct: 277 SNMVSNLSESVLIKSSLFSLMRVLSLRNCERMTLRQEVS--CSGSSSSSSGSARACSSSP 104
Query: 424 SLV 426
S+V
Sbjct: 103 SMV 95
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.315 0.129 0.343
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,780,137
Number of Sequences: 28460
Number of extensions: 80690
Number of successful extensions: 669
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 635
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 658
length of query: 495
length of database: 4,897,600
effective HSP length: 94
effective length of query: 401
effective length of database: 2,222,360
effective search space: 891166360
effective search space used: 891166360
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 57 (26.6 bits)
Medicago: description of AC133571.8


