

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC133139.5 + phase: 0
(1086 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
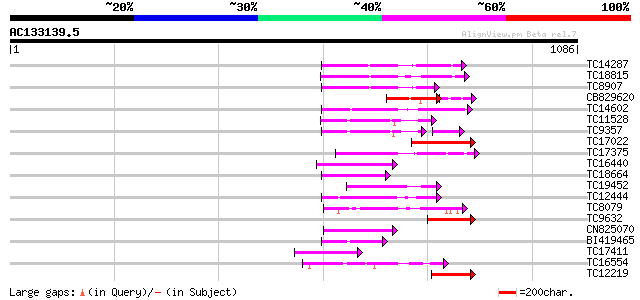
Score E
Sequences producing significant alignments: (bits) Value
TC14287 similar to UP|Q9LEU7 (Q9LEU7) Serine/threonine protein k... 151 6e-37
TC18815 similar to UP|Q8W1D5 (Q8W1D5) CBL-interacting protein ki... 147 9e-36
TC8907 similar to UP|Q8H0X3 (Q8H0X3) Serine/threonine protein ki... 127 1e-29
CB829620 94 3e-28
TC14602 UP|Q8GS56 (Q8GS56) Phosphoenolpyruvate carboxylase kinas... 120 2e-27
TC11528 homologue to UP|Q39868 (Q39868) Protein kinase , partial... 112 2e-25
TC9357 homologue to UP|Q39193 (Q39193) Protein kinase (Protein k... 103 5e-25
TC17022 homologue to UP|Q39183 (Q39183) Serine/threonine protein... 108 5e-24
TC17375 mitogen-activated kinase kinase kinase alpha [Lotus corn... 106 2e-23
TC16440 similar to UP|Q93Y18 (Q93Y18) SNF1 related protein kinas... 97 1e-20
TC18664 similar to UP|Q944I6 (Q944I6) At2g30360/T9D9.17, partial... 96 2e-20
TC19452 homologue to UP|Q8S9J9 (Q8S9J9) At1g14000/F7A19_9, parti... 96 4e-20
TC12444 similar to UP|O22981 (O22981) Ser/Thr protein kinase iso... 95 5e-20
TC8079 homologue to UP|Q9M6S1 (Q9M6S1) MAP kinase 3, complete 91 1e-18
TC9632 similar to PIR|T47546|T47546 protein kinase-like - Arabid... 88 8e-18
CN825070 84 9e-17
BI419465 82 6e-16
TC17411 similar to UP|Q8LDG6 (Q8LDG6) Protein kinase, partial (33%) 79 4e-15
TC16554 homologue to UP|AAN71719 (AAN71719) Glycogen synthase ki... 75 6e-14
TC12219 similar to UP|Q9LZS4 (Q9LZS4) Protein kinase-like protei... 74 2e-13
>TC14287 similar to UP|Q9LEU7 (Q9LEU7) Serine/threonine protein kinase-like
protein (CBL-interacting protein kinase 5), partial
(75%)
Length = 1999
Score = 151 bits (381), Expect = 6e-37
Identities = 91/278 (32%), Positives = 155/278 (55%), Gaps = 1/278 (0%)
Frame = +3
Query: 598 FEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKNAVEGILAERDILISVRNPF 657
+E+ + + +G F +V+ + +T + AIKV+KK + + V+ I E ++ VR+P
Sbjct: 432 YEMGRVLGQGNFAKVYHGRNLATNENVAIKVIKKEKLKKDRLVKQIKREVSVMRLVRHPH 611
Query: 658 VVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNLGCLDEDMARVYIAEVVLALEYLHSQSI 717
+V K ++LVMEY+ GG+L++ + N G L+ED AR Y +++ A+++ HS+ +
Sbjct: 612 IVELKEVMATKGKIFLVMEYVKGGELFTKV-NKGKLNEDDARKYFQQLISAVDFCHSRGV 788
Query: 718 VHRDLKPDNLLIGQDGHIKLTDFGLSKVGLINSTEDLSAPASFTNGFLVDDEPKPRHVSK 777
HRDLKP+NLL+ ++ +K++DFGLS + P+ R
Sbjct: 789 THRDLKPENLLLDENEDLKVSDFGLSAL------------------------PEQR---- 884
Query: 778 REARQQQSIVGTPDYLAPEILLGMGH-GTTADWWSVGVILYELLVGIPPFNADHAQQIFD 836
R+ + GTP Y+APE+L G+ G+ AD WS GVILY LL G PF ++ +I+
Sbjct: 885 RDDGMLVTPCGTPAYVAPEVLKKKGYDGSKADIWSCGVILYALLSGYLPFQGENVMRIYR 1064
Query: 837 NIINRDIQWPKHPEEISFEAYDLMNKLLIENPVQRLGV 874
+ ++ PE IS +A +L++ LL+ +P +R +
Sbjct: 1065KAFKAEYEF---PEWISPQAKNLISNLLVADPEKRYSI 1169
>TC18815 similar to UP|Q8W1D5 (Q8W1D5) CBL-interacting protein kinase
CIPK25, partial (58%)
Length = 979
Score = 147 bits (371), Expect = 9e-36
Identities = 86/287 (29%), Positives = 157/287 (53%), Gaps = 1/287 (0%)
Frame = +2
Query: 595 IEDFEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKNAVEGILAERDILISVR 654
I +E+ + + +G +V+ A++ ++G+ AIKV+ KA + ++ ++ I E I+ VR
Sbjct: 29 IGKYEMGRVLGKGTLAKVYFAKEITSGEGVAIKVMSKARIKKEGMMDQIKREISIMRLVR 208
Query: 655 NPFVVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNLGCLDEDMARVYIAEVVLALEYLHS 714
+P +V K ++ +MEY+ GG+L++ + G L +D+AR Y +++ A++Y HS
Sbjct: 209 HPNIVNLKEVMATKTKIFFIMEYIRGGELFAKVAK-GKLKDDLARRYFQQLISAVDYCHS 385
Query: 715 QSIVHRDLKPDNLLIGQDGHIKLTDFGLSKVGLINSTEDLSAPASFTNGFLVDDEPKPRH 774
+ + HRDLKP+NLL+ ++ ++K++DFGLS + P L+ +
Sbjct: 386 RGVSHRDLKPENLLLDENENLKVSDFGLSGL-----------PEQLRQDGLLHTQ----- 517
Query: 775 VSKREARQQQSIVGTPDYLAPEILLGMGH-GTTADWWSVGVILYELLVGIPPFNADHAQQ 833
GTP Y+APE+L G+ G D WS GVILY LL G PF ++
Sbjct: 518 ------------CGTPAYVAPEVLRKKGYDGFKTDTWSCGVILYALLAGCLPFQHENLMT 661
Query: 834 IFDNIINRDIQWPKHPEEISFEAYDLMNKLLIENPVQRLGVTGATEV 880
+++ ++ + Q+P S E+ L++K+L+ +P +R+ ++ V
Sbjct: 662 MYNKVLRAEFQFPPW---FSPESKKLISKILVADPNRRITISSIMRV 793
>TC8907 similar to UP|Q8H0X3 (Q8H0X3) Serine/threonine protein kinase-like
protein, partial (44%)
Length = 941
Score = 127 bits (318), Expect = 1e-29
Identities = 77/227 (33%), Positives = 123/227 (53%), Gaps = 1/227 (0%)
Frame = +3
Query: 598 FEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKNAVEGILAERDILISVRNPF 657
+EI K + +G F +V+ + T + AIKV+KK + ++ V+ I E ++ VR+P
Sbjct: 342 YEIGKILGQGNFAKVYHGRNMETNESVAIKVIKKERLKKERLVKQIKREVSVMRLVRHPH 521
Query: 658 VVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNLGCLDEDMARVYIAEVVLALEYLHSQSI 717
+V K +++V+EY+ GG+L++ L G + E AR Y +++ A+++ HS+ +
Sbjct: 522 IVELKEVMATKTKIFMVVEYVKGGELFAKLTK-GKMTEVAARKYFQQLISAVDFCHSRGV 698
Query: 718 VHRDLKPDNLLIGQDGHIKLTDFGLSKVGLINSTEDLSAPASFTNGFLVDDEPKPRHVSK 777
HRDLKP+NLL+ + +K++DFGLS + P+ R
Sbjct: 699 THRDLKPENLLLDDNEDLKVSDFGLSSL------------------------PEQR---- 794
Query: 778 REARQQQSIVGTPDYLAPEILLGMGH-GTTADWWSVGVILYELLVGI 823
R + GTP Y+APE+L G+ G+ AD WS GVILY LL GI
Sbjct: 795 RSDGMLLTPCGTPAYVAPEVLKKKGYDGSKADIWSCGVILYALLCGI 935
>CB829620
Length = 548
Score = 93.6 bits (231), Expect(2) = 3e-28
Identities = 52/118 (44%), Positives = 72/118 (60%), Gaps = 14/118 (11%)
Frame = +1
Query: 723 KPDNLLIGQDGHIKLTDFGLSK-VGLINSTEDLSAPASFTNGFLVDDEPKPRHVSKREAR 781
KPDNLL+ ++GH+KL+DFGL K + N E+ + S +G L + R V+ + +
Sbjct: 1 KPDNLLLDRNGHMKLSDFGLCKPLDCSNLQENDFSTGSNRSGAL---QSNGRPVAPKRTQ 171
Query: 782 QQQ-------------SIVGTPDYLAPEILLGMGHGTTADWWSVGVILYELLVGIPPF 826
Q+Q S VGTPDY+APE+LL G+G DWWS+G I+YE+LVG PPF
Sbjct: 172 QEQLQHWQKNRRTLAYSTVGTPDYIAPEVLLKKGYGMECDWWSLGGIMYEMLVGYPPF 345
Score = 50.1 bits (118), Expect(2) = 3e-28
Identities = 33/77 (42%), Positives = 42/77 (53%), Gaps = 5/77 (6%)
Frame = +3
Query: 823 IPPFNADHAQQIFDNIINRDIQWPKH---PEE--ISFEAYDLMNKLLIENPVQRLGVTGA 877
I PF +D I+N W H PEE +S EA DL+++LL N QRLG GA
Sbjct: 333 ISPFYSDEPMLTCRKIVN----WRTHLKFPEEAKLSPEAKDLISRLLC-NVQQRLGTKGA 497
Query: 878 TEVKRHAFFKDVNWDTL 894
E+K H +FK + WD L
Sbjct: 498 DEIKAHPWFKGIEWDKL 548
>TC14602 UP|Q8GS56 (Q8GS56) Phosphoenolpyruvate carboxylase kinase, complete
Length = 1256
Score = 120 bits (300), Expect = 2e-27
Identities = 85/295 (28%), Positives = 143/295 (47%), Gaps = 6/295 (2%)
Frame = +2
Query: 598 FEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKK---ADMIRKNAVEGILAERDILISVR 654
+++ + I RG FG +F + D FA+K++ K AD ++ +E +L
Sbjct: 68 YQLCEEIGRGRFGTIFRCFHPISTDTFAVKLIDKSLLADSTDRHCLENEPKYMSLLSP-- 241
Query: 655 NPFVVRFYYSFTCKENLYLVMEYLNGGDLYSML--RNLGCLDEDMARVYIAEVVLALEYL 712
+P +++ + F + L +V+E L + N + E A + +++ A+ +
Sbjct: 242 HPNILQIFDVFEDDDVLSMVIELCQPLTLLDRIVAANGTSIPEVEAAGLMKQLLEAVAHC 421
Query: 713 HSQSIVHRDLKPDNLLIGQDGHIKLTDFGLSKVGLINSTEDLSAPASFTNGFLVDDEPKP 772
H + HRD+KPDN+L G G +KL DFG ++ F +G
Sbjct: 422 HRLGVAHRDVKPDNVLFGGGGDLKLADFGSAEW--------------FGDG--------- 532
Query: 773 RHVSKREARQQQSIVGTPDYLAPEILLGMGHGTTADWWSVGVILYELLVGIPPFNADHAQ 832
R+ +VGTP Y+APE+L+G +G D WS GVILY +L G PPF D A
Sbjct: 533 --------RRMSGVVGTPYYVAPEVLMGREYGEKVDVWSCGVILYIMLSGTPPFYGDSAA 688
Query: 833 QIFDNIINRDIQWPKHP-EEISFEAYDLMNKLLIENPVQRLGVTGATEVKRHAFF 886
+IF+ +I ++++P +S A DL+ K++ +P R+ A + RH +F
Sbjct: 689 EIFEAVIRGNLRFPSRIFRNVSPAAKDLLRKMICRDPSNRI---SAEQALRHPWF 844
>TC11528 homologue to UP|Q39868 (Q39868) Protein kinase , partial (56%)
Length = 640
Score = 112 bits (281), Expect = 2e-25
Identities = 73/226 (32%), Positives = 115/226 (50%), Gaps = 5/226 (2%)
Frame = +3
Query: 596 EDFEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKNAVEGILAERDILISVRN 655
E +E +K + G FG LA+ ++TG+L A+K +++ I +N I+ R S+R+
Sbjct: 72 ERYEPLKELGSGNFGVARLARDKNTGELVAVKYIERGKKIDENVQREIINHR----SLRH 239
Query: 656 PFVVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNLGCLDEDMARVYIAEVVLALEYLHSQ 715
P ++RF +L +V+EY +GG+L+ + + G ED AR + +++ + Y HS
Sbjct: 240 PNIIRFKEVLLTPTHLAIVLEYASGGELFERICSAGRFSEDEARYFFQQLISGVSYCHSM 419
Query: 716 SIVHRDLKPDNLLIGQDGH----IKLTDFGLSKVGLINSTEDLSAPASFTNGFLVDDEPK 771
I HRDLK +N L+ DG+ +K+ DFG SK +++S
Sbjct: 420 EICHRDLKLENTLL--DGNPSPRLKICDFGYSKSAILHS--------------------- 530
Query: 772 PRHVSKREARQQQSIVGTPDYLAPEILLGMGH-GTTADWWSVGVIL 816
Q +S VGTP Y+APE+L + G AD WS GV L
Sbjct: 531 ----------QPKSTVGTPAYIAPEVLSRKEYDGKVADVWSCGVTL 638
>TC9357 homologue to UP|Q39193 (Q39193) Protein kinase (Protein kinase,
41K) , partial (71%)
Length = 1181
Score = 103 bits (257), Expect(2) = 5e-25
Identities = 64/205 (31%), Positives = 103/205 (50%), Gaps = 4/205 (1%)
Frame = +1
Query: 598 FEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKNAVEGILAERDILISVRNPF 657
+++++ I G FG L Q + T +L A+K +++ D I +N I+ R S+R+P
Sbjct: 361 YDLVRDIGSGNFGVARLMQDKQTKELVAVKYIERGDKIDENVKREIINHR----SLRHPN 528
Query: 658 VVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNLGCLDEDMARVYIAEVVLALEYLHSQSI 717
+VRF +L +VMEY +GG+L+ + N G ED AR + +++ + Y H+ +
Sbjct: 529 IVRFKEVILTPTHLAIVMEYASGGELFERICNAGRFTEDEARFFFQQLISGVSYCHAMQV 708
Query: 718 VHRDLKPDNLLIGQDG----HIKLTDFGLSKVGLINSTEDLSAPASFTNGFLVDDEPKPR 773
HRDLK +N L+ DG +K+ DFG SK +++S
Sbjct: 709 CHRDLKLENTLL--DGSPTPRLKICDFGYSKSSVLHS----------------------- 813
Query: 774 HVSKREARQQQSIVGTPDYLAPEIL 798
Q +S VGTP Y+APE+L
Sbjct: 814 --------QPKSTVGTPAYIAPEVL 864
Score = 28.9 bits (63), Expect(2) = 5e-25
Identities = 21/65 (32%), Positives = 35/65 (53%), Gaps = 4/65 (6%)
Frame = +3
Query: 811 SVGVILYELLVGIPPFNADHAQQIFDNIINR--DIQWPKHPE--EISFEAYDLMNKLLIE 866
S GV LY +LVG PF + + F I R +Q+ P+ +IS E L++++ +
Sbjct: 906 SCGVTLYVMLVGAYPFEDPNEPKDFRKTIQRVLSVQY-SIPDFVQISPECRHLISRIFVF 1082
Query: 867 NPVQR 871
+P +R
Sbjct: 1083DPAER 1097
>TC17022 homologue to UP|Q39183 (Q39183) Serine/threonine protein kinase
(Protein kinase 5) (AT5G47750/MCA23_7) , partial (32%)
Length = 775
Score = 108 bits (270), Expect = 5e-24
Identities = 52/123 (42%), Positives = 76/123 (61%), Gaps = 1/123 (0%)
Frame = +1
Query: 770 PKPRHVSKREARQQQSIVGTPDYLAPEILLGMGHGTTADWWSVGVILYELLVGIPPFNAD 829
P P +++ + S VGT +YLAPEI+ G GHG+ DWW+ G+ LYELL G PF
Sbjct: 103 PLPELIAEPTDARSMSFVGTHEYLAPEIIKGEGHGSAVDWWTFGIFLYELLFGKTPFKGS 282
Query: 830 HAQQIFDNIINRDIQWPKHPEEISFEAYDLMNKLLIENPVQRLGV-TGATEVKRHAFFKD 888
+ N++ + +++P+ P +SF A DL+ LL++ P RL GATE+K+H FF+
Sbjct: 283 GNRATLFNVVGQPLRFPEAP-VVSFAARDLIRGLLVKEPQHRLAYKRGATEIKQHPFFEG 459
Query: 889 VNW 891
VNW
Sbjct: 460 VNW 468
>TC17375 mitogen-activated kinase kinase kinase alpha [Lotus corniculatus
var. japonicus]
Length = 1422
Score = 106 bits (265), Expect = 2e-23
Identities = 76/279 (27%), Positives = 135/279 (48%), Gaps = 4/279 (1%)
Frame = +3
Query: 625 AIKVLK--KADMIRKNAVEGILAERDILISVRNPFVVRFYYSFTCKENLYLVMEYLNGGD 682
AIK +K D K ++ + E ++L +P +V++Y S +E+L + +EY++GG
Sbjct: 12 AIKEVKVFSDDKTSKECLKQLNQEINLLNQFSHPNIVQYYGSELGEESLSVYLEYVSGGS 191
Query: 683 LYSMLRNLGCLDEDMARVYIAEVVLALEYLHSQSIVHRDLKPDNLLIGQDGHIKLTDFGL 742
++ +L+ G E + + Y ++V L YLHS++ VHRD+K N+L+ +G IKL DFG+
Sbjct: 192 IHKLLQEYGAFKEPVIQNYTRQIVSGLAYLHSRNTVHRDIKGANILVDPNGEIKLADFGM 371
Query: 743 SKVGLINSTEDLSAPASFTNGFLVDDEPKPRHVSKREARQQQSIVGTPDYLAPEILLGM- 801
SK H++ A S G+P ++APE+++
Sbjct: 372 SK-----------------------------HIN--SAASMLSFKGSPYWMAPEVVMNTN 458
Query: 802 GHGTTADWWSVGVILYELLVGIPPFNA-DHAQQIFDNIINRDIQWPKHPEEISFEAYDLM 860
G+G D S+G + E+ PP++ + IF I P+ PE +S +A + +
Sbjct: 459 GYGLPVDISSLGCTILEMATSKPPWSQFEGVAAIFK--IGNSKDMPEIPEHLSDDAKNFI 632
Query: 861 NKLLIENPVQRLGVTGATEVKRHAFFKDVNWDTLARQKV 899
+ L +P+ R A + H F +D + +A +
Sbjct: 633 KQCLQRDPLAR---PTAQSLLNHPFIRDQSATKVANASI 740
>TC16440 similar to UP|Q93Y18 (Q93Y18) SNF1 related protein kinase, partial
(30%)
Length = 549
Score = 97.4 bits (241), Expect = 1e-20
Identities = 48/156 (30%), Positives = 91/156 (57%), Gaps = 1/156 (0%)
Frame = +1
Query: 589 SKDRTSIEDFEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKNAVEGILAERD 648
S R + +++ + + RG F +V+ A + G A+K++ K+ + + I+ E D
Sbjct: 64 SPPRIILGKYQLTRFLGRGNFAKVYQAVSLTDGTTVAVKMIDKSKTVDASMEPRIVREID 243
Query: 649 ILISVRN-PFVVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNLGCLDEDMARVYIAEVVL 707
+ +++ P +++ + + +YL+++Y GG+L+S + G L E +AR Y ++V
Sbjct: 244 AMRRLQHHPNILKIHEVMATRTKIYLIVDYAGGGELFSKISRRGRLPEPLARRYFQQLVS 423
Query: 708 ALEYLHSQSIVHRDLKPDNLLIGQDGHIKLTDFGLS 743
AL + H + HRDLKP NLL+ +G++K++DFGLS
Sbjct: 424 ALCFCHRNGVAHRDLKPQNLLLDAEGNLKVSDFGLS 531
>TC18664 similar to UP|Q944I6 (Q944I6) At2g30360/T9D9.17, partial (34%)
Length = 643
Score = 96.3 bits (238), Expect = 2e-20
Identities = 46/132 (34%), Positives = 77/132 (57%)
Frame = +3
Query: 598 FEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKNAVEGILAERDILISVRNPF 657
+E+ + + GAF +V+ A+ TG A+KV+ K +I + E I+ +R+P
Sbjct: 93 YEVGRLLGCGAFAKVYHARNIETGQSVAVKVINKKKVIGTGLTGHVKREVSIMSRLRHPN 272
Query: 658 VVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNLGCLDEDMARVYIAEVVLALEYLHSQSI 717
+VR + K +Y VME+ GG+L++ + G ED+AR Y +++ A+ Y HS+ +
Sbjct: 273 IVRLHEVLATKTKIYFVMEFAKGGELFARISTKGRFSEDLARRYFQQLISAVGYCHSRGV 452
Query: 718 VHRDLKPDNLLI 729
HRDLKP+NLL+
Sbjct: 453 FHRDLKPENLLL 488
>TC19452 homologue to UP|Q8S9J9 (Q8S9J9) At1g14000/F7A19_9, partial (41%)
Length = 562
Score = 95.5 bits (236), Expect = 4e-20
Identities = 59/185 (31%), Positives = 95/185 (50%), Gaps = 4/185 (2%)
Frame = +3
Query: 646 ERDILISVRNPFVVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNLGCLDEDMARVYIAEV 705
E ++L+ +R+P +V+F + T ++ L L+ EYL GGDL+ L+ G L A + ++
Sbjct: 48 EVNLLVKLRHPNIVQFLGAVTERKPLMLITEYLRGGDLHQYLKEKGSLSPSTAINFSMDI 227
Query: 706 VLALEYLHSQS--IVHRDLKPDNLLIGQDG--HIKLTDFGLSKVGLINSTEDLSAPASFT 761
V + YLH++ I+HRDLKP N+L+ H+K+ DFGLSK+ + ++ D+ T
Sbjct: 228 VRGMAYLHNEPNVIIHRDLKPRNVLLVNSSADHLKVGDFGLSKLITVQNSHDVYKMTGET 407
Query: 762 NGFLVDDEPKPRHVSKREARQQQSIVGTPDYLAPEILLGMGHGTTADWWSVGVILYELLV 821
G+ Y+APE+ + D +S +ILYE+L
Sbjct: 408 --------------------------GSYRYMAPEVFKHRKYDKKVDVYSFAMILYEMLE 509
Query: 822 GIPPF 826
G PPF
Sbjct: 510 GEPPF 524
>TC12444 similar to UP|O22981 (O22981) Ser/Thr protein kinase isolog,
partial (37%)
Length = 709
Score = 95.1 bits (235), Expect = 5e-20
Identities = 69/235 (29%), Positives = 111/235 (46%), Gaps = 4/235 (1%)
Frame = +2
Query: 597 DFEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKNA-VEGILAERDILISVRN 655
D+ +++ + GA V+ A + A+K + D+ R NA +E I E + + +
Sbjct: 86 DYNLLEEVGYGASATVYRAVFLPRDEEVAVKCV---DLDRCNANLEDIRREAQTMSLIDH 256
Query: 656 PFVVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNL--GCLDEDMARVYIAEVVLALEYLH 713
VVR ++SF L++VM ++ G +++ +E+ + E + ALEYLH
Sbjct: 257 RNVVRAHWSFVVDRRLWVVMPFMAQGSCLHLMKVAYPDGFEEEAIGSILKETLKALEYLH 436
Query: 714 SQSIVHRDLKPDNLLIGQDGHIKLTDFGLSKVGLINSTEDLSAPASFTNGFLVDDEPKPR 773
+HRD+K N+L+G DG +KL DFG+S + F G
Sbjct: 437 RHGHIHRDVKAGNILLGSDGQVKLADFGVS-------------ASMFDAG---------- 547
Query: 774 HVSKREARQQQSIVGTPDYLAPEILL-GMGHGTTADWWSVGVILYELLVGIPPFN 827
R + + VGTP ++APE+L G G+ AD WS G+ EL G PF+
Sbjct: 548 ----ERQRNRNTFVGTPCWMAPEVLQPGTGYNFKADVWSFGITALELAHGHAPFS 700
>TC8079 homologue to UP|Q9M6S1 (Q9M6S1) MAP kinase 3, complete
Length = 1550
Score = 90.9 bits (224), Expect = 1e-18
Identities = 87/320 (27%), Positives = 143/320 (44%), Gaps = 43/320 (13%)
Frame = +3
Query: 601 IKPISRGAFGRVFLAQKRSTGDLFAIK--------------VLKKADMIRKNAVEGILAE 646
I PI RGA+G V T +L A+K L++ ++R E ++A
Sbjct: 300 IMPIGRGAYGIVCSLLNTETNELVAVKKIANAFDNHMDAKRTLREIKLLRHLDHENVIAL 479
Query: 647 RDILISVRNPFVVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNLGCLDEDMARVYIAEVV 706
RD++ P +R ++ ++Y+ E ++ DL+ ++R+ L E+ + ++ +V+
Sbjct: 480 RDVI-----PPPLRREFT-----DVYITTELMDT-DLHQIIRSNQGLSEEHCQYFLYQVL 626
Query: 707 LALEYLHSQSIVHRDLKPDNLLIGQDGHIKLTDFGLSKVGLINSTEDLSAPASFTNGFLV 766
L+Y+HS +I+HRDLKP NLL+ + +K+ DFGL++ + N F+
Sbjct: 627 RGLKYIHSANIIHRDLKPSNLLLNSNCDLKIIDFGLAR-------------PTVENDFMT 767
Query: 767 DDEPKPRHVSKREARQQQSIVGTPDYLAPEILLGMGHGTTA-DWWSVGVILYELLVGIPP 825
+ V T Y APE+LL T+A D WSVG I EL+ P
Sbjct: 768 E------------------YVVTRWYRAPELLLNSSDYTSAIDVWSVGCIFMELMNKKPL 893
Query: 826 F-NADHAQQI--------------FDNIINRDI--------QWPKHPEEISFE-----AY 857
F DH Q+ + N D+ Q+P+ P F A
Sbjct: 894 FPGKDHVHQLRLLTELLGTPTEADLGLVKNDDVRRYIRQLPQYPRQPLTKVFPHVHPMAM 1073
Query: 858 DLMNKLLIENPVQRLGVTGA 877
DL++K+L +P +R+ V A
Sbjct: 1074DLVDKMLTIDPTKRITVEQA 1133
>TC9632 similar to PIR|T47546|T47546 protein kinase-like - Arabidopsis
thaliana {Arabidopsis thaliana;}, partial (12%)
Length = 650
Score = 87.8 bits (216), Expect = 8e-18
Identities = 40/93 (43%), Positives = 59/93 (63%), Gaps = 1/93 (1%)
Frame = +3
Query: 800 GMGHGTTADWWSVGVILYELLVGIPPFNADHAQQIFDNIINRDIQWPKHPEEISFEAYDL 859
G GHG DWW+ GV LYELL G PF + ++ N++ + +++P P +SF+A DL
Sbjct: 6 GEGHGAAVDWWTFGVFLYELLYGRTPFKGSNNEETLANVVLQGLRFPDSP-FVSFQARDL 182
Query: 860 MNKLLIENPVQRLGV-TGATEVKRHAFFKDVNW 891
+ LL++ P RLG GA E+K+H FF+ +NW
Sbjct: 183 IRGLLVKEPENRLGTEKGAAEIKQHPFFEGLNW 281
>CN825070
Length = 634
Score = 84.3 bits (207), Expect = 9e-17
Identities = 48/146 (32%), Positives = 79/146 (53%), Gaps = 4/146 (2%)
Frame = +3
Query: 602 KPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKNAVEGILAERDILISV-RNPFVVR 660
+ + +G FG +L + STG FA K + K ++ K + + E I+ + +P VVR
Sbjct: 141 RKLGQGQFGTTYLCRHNSTGCTFACKSIPKRKLLCKEDYDDVWREIQIMHHLSEHPHVVR 320
Query: 661 FYYSFTCKENLYLVMEYLNGGDLYSMLRNLGCLDEDMARVYIAEVVLALEYLHSQSIVHR 720
+ ++ ++++VME GG+L+ + G E A I +V +E HS ++HR
Sbjct: 321 IHGTYEDAASVHIVMEICEGGELFDRIVQKGQYSEREAAKLIRTIVEVVEACHSLGVMHR 500
Query: 721 DLKPDNLL---IGQDGHIKLTDFGLS 743
DLKP+N L + +D +K TDFGLS
Sbjct: 501 DLKPENFLFDTVEEDAKLKTTDFGLS 578
>BI419465
Length = 487
Score = 81.6 bits (200), Expect = 6e-16
Identities = 42/126 (33%), Positives = 70/126 (55%)
Frame = +3
Query: 598 FEIIKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKNAVEGILAERDILISVRNPF 657
++ ++ I G FG L + + T +L A+K +++ D I +N I+ R S+R+P
Sbjct: 114 YDFVRDIGSGNFGVARLMRDKHTKELVAVKYIERGDKIYENVKREIINHR----SLRHPN 281
Query: 658 VVRFYYSFTCKENLYLVMEYLNGGDLYSMLRNLGCLDEDMARVYIAEVVLALEYLHSQSI 717
+VRF +L +VMEY +GG+L+ + + G ED AR + +++ + Y HS I
Sbjct: 282 IVRFKEVILTPTHLAIVMEYASGGELFEKIHSAGRFTEDEARFFFQQLISGVSYCHSMQI 461
Query: 718 VHRDLK 723
HRDLK
Sbjct: 462 CHRDLK 479
>TC17411 similar to UP|Q8LDG6 (Q8LDG6) Protein kinase, partial (33%)
Length = 632
Score = 79.0 bits (193), Expect = 4e-15
Identities = 44/136 (32%), Positives = 78/136 (57%), Gaps = 4/136 (2%)
Frame = +2
Query: 545 EKYLTLCEQIHDERAESSNSMADEESSVDD--DTIRSL--RASPINGFSKDRTSIEDFEI 600
+ YL + D R + + + S ++ + +R+L R + + + I+DFE
Sbjct: 224 KNYLQGLQDRKDRRRALQRKVQESQVSAEEQEEMMRNLARRETEYMRLQRRKIGIDDFEQ 403
Query: 601 IKPISRGAFGRVFLAQKRSTGDLFAIKVLKKADMIRKNAVEGILAERDILISVRNPFVVR 660
+ I +GAFG V L + + TG++FA+K LKK+DM+ + VE + +ER++L V + +V+
Sbjct: 404 LTVIGKGAFGEVRLCRAKDTGEIFAMKKLKKSDMLSRGQVEHVRSERNLLAEVDSRCIVK 583
Query: 661 FYYSFTCKENLYLVME 676
+YSF + LYL+ME
Sbjct: 584 LHYSFQDSDFLYLIME 631
>TC16554 homologue to UP|AAN71719 (AAN71719) Glycogen synthase kinase 3 beta
protein kinase DWARF12 , partial (71%)
Length = 927
Score = 75.1 bits (183), Expect = 6e-14
Identities = 79/295 (26%), Positives = 135/295 (44%), Gaps = 17/295 (5%)
Frame = +1
Query: 562 SNSMA-DEESSV-----DDDTIRSLRASPINGFSKDRTSIEDFEIIKPISRGAFGRVFLA 615
S+SMA D+E S +D + ++ I G + + + + + G+FG VF A
Sbjct: 112 SSSMAEDKEMSTSVINGNDSLTGHIISTTIGGKNGEPKQTISYMAERVVGTGSFGIVFQA 291
Query: 616 QKRSTGDLFAIKVLKKADMIRKNAVEGILAERDILISVRNPFVVRF---YYSFTCKENLY 672
+ +G+ AIK + + D KN E ++ + +P VV ++S T + L+
Sbjct: 292 KCLESGEAVAIKKVLQ-DRRYKNR------ELQLMRVMDHPNVVSLKHCFFSTTSTDELF 450
Query: 673 L--VMEYLNGGDLYSMLRNLGCLDEDM----ARVYIAEVVLALEYLHS-QSIVHRDLKPD 725
L VMEY+ +Y ++++ ++ M ++Y+ ++ L Y+H+ + HRDLKP
Sbjct: 451 LNLVMEYVPES-MYRVIKHYSNANQRMPIIYVKLYMYQIFRGLAYIHTVPGVCHRDLKPQ 627
Query: 726 NLLIGQDGH-IKLTDFGLSKVGLINSTEDLSAPASFTNGFLVDDEPKPRHVSKREARQQQ 784
N+L+ H +KL DFG +K+ LV E ++ R R +
Sbjct: 628 NILVDPLTHQVKLCDFGSAKM-------------------LVKGEANISYICSRFYRAPE 750
Query: 785 SIVGTPDYLAPEILLGMGHGTTADWWSVGVILYELLVGIPPFNADHAQQIFDNII 839
I G +Y T+ D WS G +L ELL+G P F ++A +II
Sbjct: 751 LIFGATEY-----------STSIDIWSAGCVLAELLLGQPLFPGENAVDQLVHII 882
>TC12219 similar to UP|Q9LZS4 (Q9LZS4) Protein kinase-like protein, partial
(11%)
Length = 587
Score = 73.6 bits (179), Expect = 2e-13
Identities = 35/85 (41%), Positives = 54/85 (63%), Gaps = 1/85 (1%)
Frame = +3
Query: 808 DWWSVGVILYELLVGIPPFNADHAQQIFDNIINRDIQWPKHPEEISFEAYDLMNKLLIEN 867
DWW+ G+ L+ELL G PF + N++++ +++P P +SF A DL+ LLI++
Sbjct: 12 DWWTFGIFLFELLYGKTPFKGLANEDTLANVVSQSLKFPSAP-IVSFHARDLIRGLLIKD 188
Query: 868 PVQRLG-VTGATEVKRHAFFKDVNW 891
P RLG V GA E+K+H F + +NW
Sbjct: 189 PENRLGSVKGAAEIKQHPFSEGLNW 263
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.317 0.134 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,661,212
Number of Sequences: 28460
Number of extensions: 239850
Number of successful extensions: 1502
Number of sequences better than 10.0: 197
Number of HSP's better than 10.0 without gapping: 1353
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1451
length of query: 1086
length of database: 4,897,600
effective HSP length: 100
effective length of query: 986
effective length of database: 2,051,600
effective search space: 2022877600
effective search space used: 2022877600
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC133139.5


