

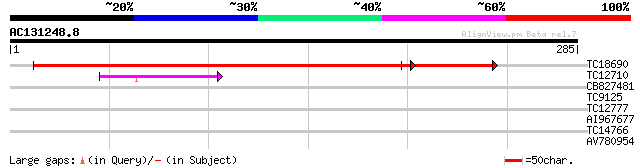
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC131248.8 + phase: 0
(285 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC18690 similar to UP|P93470 (P93470) Cholinephosphate cytidylyl... 348 e-110
TC12710 similar to UP|Q84X92 (Q84X92) Phosphoethanolamine cytidy... 43 5e-05
CB827481 38 0.002
TC9125 similar to UP|Q40316 (Q40316) Vestitone reductase, complete 27 3.0
TC12777 similar to UP|Q9SYP6 (Q9SYP6) F9H16.10 protein, partial ... 27 5.2
AI967677 26 6.8
TC14766 similar to UP|Q7XXR8 (Q7XXR8) Nascent polypeptide associ... 26 6.8
AV780954 26 8.9
>TC18690 similar to UP|P93470 (P93470) Cholinephosphate cytidylyltransferase
, partial (80%)
Length = 845
Score = 348 bits (894), Expect(2) = e-110
Identities = 163/192 (84%), Positives = 179/192 (92%)
Frame = +2
Query: 13 DKPVRVYADGIYDLFHFGHARSLEQAKKSFPNTYLLVGCCSDEITHKYKGKTVMNEQERY 72
D+PVRVYADGIYDLFHFGHA SLEQAKKSFPNTYLLVGCC+DE+THKYKGKTVM E ERY
Sbjct: 77 DRPVRVYADGIYDLFHFGHAGSLEQAKKSFPNTYLLVGCCNDEVTHKYKGKTVMTEAERY 256
Query: 73 ESLRHCKWVDEVIPDVPWVINQEFIDKHKIDYVAHDALPYADTSGAGNDVYEFVKAIGKF 132
ESLRHCKWVDEVIPD PWVINQEF+DKH+IDYVAHDALPYADTSGA NDVYEFVKA+GKF
Sbjct: 257 ESLRHCKWVDEVIPDAPWVINQEFLDKHRIDYVAHDALPYADTSGAANDVYEFVKAVGKF 436
Query: 133 KETKRTEGISTSDIIMRIIKDYNQYVMRNLDRGYSRKELGVSYVKEKRLRMNMGLKKLQE 192
KET+RT+GISTSDIIMRI+KDYNQYV+RNLDRGYSRKELGVSYVKEKRLR+N LK L E
Sbjct: 437 KETQRTDGISTSDIIMRIVKDYNQYVLRNLDRGYSRKELGVSYVKEKRLRVNXRLKTLHE 616
Query: 193 RVKKQQETVGKK 204
+VK+ ++ K+
Sbjct: 617 KVKEHKKRXVKR 652
Score = 65.5 bits (158), Expect(2) = e-110
Identities = 30/48 (62%), Positives = 36/48 (74%)
Frame = +1
Query: 198 QETVGKKIGTVRRIAGMNRTEWVENADRLVAGFLEMFEEGCHKMGTAI 245
QE VG+KI V AG +R EWVENAD+ VAG LEMFE+GCH GT++
Sbjct: 631 QEKVGEKIQIVAXTAGXHRNEWVENADKWVAGILEMFEKGCHXXGTSL 774
>TC12710 similar to UP|Q84X92 (Q84X92) Phosphoethanolamine
cytidylyltransferase, partial (33%)
Length = 515
Score = 43.1 bits (100), Expect = 5e-05
Identities = 22/64 (34%), Positives = 36/64 (55%), Gaps = 2/64 (3%)
Frame = +3
Query: 46 YLLVGCCSDEITHKYKG--KTVMNEQERYESLRHCKWVDEVIPDVPWVINQEFIDKHKID 103
+LLVG +D + +G + +M+ ER S+ C++VDEVI PW ++++ I I
Sbjct: 9 FLLVGIHTDHTVSETRGLHRPIMHLHERSLSVLACRYVDEVIIGAPWEVSKDMITTFNIS 188
Query: 104 YVAH 107
V H
Sbjct: 189 LVVH 200
>CB827481
Length = 556
Score = 38.1 bits (87), Expect = 0.002
Identities = 19/38 (50%), Positives = 26/38 (68%)
Frame = +2
Query: 13 DKPVRVYADGIYDLFHFGHARSLEQAKKSFPNTYLLVG 50
D+P RVY GIYD+F+FG SL+QAK S+ + +G
Sbjct: 68 DRPDRVY--GIYDVFNFGDVASLKQAKNSYSSVVEKMG 175
>TC9125 similar to UP|Q40316 (Q40316) Vestitone reductase, complete
Length = 1200
Score = 27.3 bits (59), Expect = 3.0
Identities = 19/54 (35%), Positives = 26/54 (47%)
Frame = -3
Query: 14 KPVRVYADGIYDLFHFGHARSLEQAKKSFPNTYLLVGCCSDEITHKYKGKTVMN 67
+P+ VY + +F F HA + +A P+T LLV S T K G V N
Sbjct: 433 EPLEVYTTR-FTVFEFTHAFKMPRA----PSTVLLVTTSSGSFTAKSTGVAVWN 287
>TC12777 similar to UP|Q9SYP6 (Q9SYP6) F9H16.10 protein, partial (10%)
Length = 500
Score = 26.6 bits (57), Expect = 5.2
Identities = 15/58 (25%), Positives = 31/58 (52%), Gaps = 3/58 (5%)
Frame = +1
Query: 151 IKDYNQ---YVMRNLDRGYSRKELGVSYVKEKRLRMNMGLKKLQERVKKQQETVGKKI 205
IKD+++ VM + ++++ + KR NMGLKK + +K + +G+++
Sbjct: 175 IKDWSKRMGLVMEEVALSLMKRKMR*GKQQRKRKPKNMGLKKTSQILKMKMMVLGRQV 348
>AI967677
Length = 407
Score = 26.2 bits (56), Expect = 6.8
Identities = 12/28 (42%), Positives = 18/28 (63%)
Frame = +1
Query: 111 PYADTSGAGNDVYEFVKAIGKFKETKRT 138
P DTS AGN++Y +K + K+ KR+
Sbjct: 139 PGDDTSAAGNNLYGNLKQLYLLKQRKRS 222
>TC14766 similar to UP|Q7XXR8 (Q7XXR8) Nascent polypeptide associated
complex alpha chain, partial (58%)
Length = 600
Score = 26.2 bits (56), Expect = 6.8
Identities = 18/55 (32%), Positives = 28/55 (50%), Gaps = 10/55 (18%)
Frame = +2
Query: 237 GCHKMGTAIRDRIQEQLKAQQLKSLLY----------DEWDDDVDDEFYEDESVE 281
G H+ TA+ + QE+L A L+ DE DDD DD+ +D+++E
Sbjct: 38 GTHR-STAMTAQSQEELLAAHLEQQKIHDDEPVVEDDDEDDDDEDDDDEDDDNIE 199
>AV780954
Length = 476
Score = 25.8 bits (55), Expect = 8.9
Identities = 13/32 (40%), Positives = 20/32 (61%)
Frame = +2
Query: 151 IKDYNQYVMRNLDRGYSRKELGVSYVKEKRLR 182
I+DYN+ MR + YSRK +G+S K + +
Sbjct: 368 IEDYNE--MRKYNG*YSRKVIGISKTKTQEAK 457
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.317 0.136 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,024,585
Number of Sequences: 28460
Number of extensions: 46122
Number of successful extensions: 255
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 249
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 255
length of query: 285
length of database: 4,897,600
effective HSP length: 89
effective length of query: 196
effective length of database: 2,364,660
effective search space: 463473360
effective search space used: 463473360
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 55 (25.8 bits)
Medicago: description of AC131248.8


