

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC130806.14 - phase: 0
(394 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
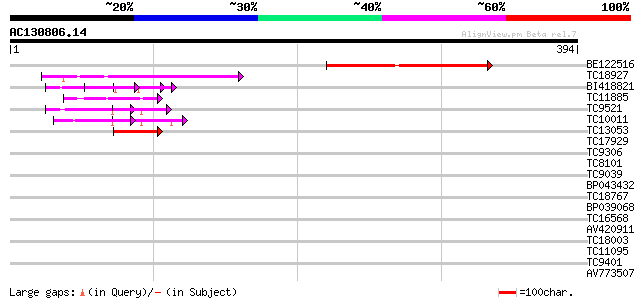
Score E
Sequences producing significant alignments: (bits) Value
BE122516 91 3e-19
TC18927 similar to PIR|AI2934|AI2934 chromate transport protein ... 71 4e-13
BI418821 57 4e-09
TC11885 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, parti... 46 1e-05
TC9521 similar to UP|Q9FYA7 (Q9FYA7) Splicing factor RSZ33, part... 45 2e-05
TC10011 similar to UP|Q9FYA7 (Q9FYA7) Splicing factor RSZ33, par... 45 2e-05
TC13053 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, parti... 42 2e-04
TC17929 32 0.14
TC9306 similar to UP|Q42922 (Q42922) Annexin (Fragment), partial... 30 0.70
TC8101 similar to UP|O24074 (O24074) DNAJ-like protein, partial ... 29 1.2
TC9039 29 1.6
BP043432 28 2.0
TC18767 28 2.6
BP039068 28 2.6
TC16568 similar to UP|AAR82759 (AAR82759) RE40656p (Fragment), p... 28 2.6
AV420911 27 4.5
TC18003 similar to PIR|T05112|T05112 splicing factor 9G8-like SR... 27 4.5
TC11095 similar to PIR|T05112|T05112 splicing factor 9G8-like SR... 27 5.9
TC9401 similar to UP|Q84LW7 (Q84LW7) Peptidylprolyl cis-trans is... 27 5.9
AV773507 27 7.7
>BE122516
Length = 364
Score = 91.3 bits (225), Expect = 3e-19
Identities = 51/115 (44%), Positives = 73/115 (63%)
Frame = +2
Query: 221 GMNWLEYNRVHINCFNKTVHFSSAEEESGVEILSTKEMKQLERDGILMYSLMACLSLENQ 280
GMNWL N +NC KTV F ++E ++ + K K E + ++ L A + ++
Sbjct: 17 GMNWLTANDATLNCRKKTVTFGTSEGDAKRVKRTDKVGKASECESDVL--LGALETDKSD 190
Query: 281 AVIDKLPVVNEFPEVFSDEIPDVPPEREVEFSIDLVPRTKPVSMAPYRMSASELA 335
++ +PVV EF +VF +E+ ++PPEREVEFSID VP T P+S+APYRMS ELA
Sbjct: 191 TGVEGIPVVREFSDVFPEEVSELPPEREVEFSID*VPGTGPISIAPYRMSLVELA 355
>TC18927 similar to PIR|AI2934|AI2934 chromate transport protein chrA
[imported] - Agrobacterium tumefaciens
(strain C58, Dupont) {Agrobacterium tumefaciens;},
partial (6%)
Length = 561
Score = 70.9 bits (172), Expect = 4e-13
Identities = 46/147 (31%), Positives = 67/147 (45%), Gaps = 7/147 (4%)
Frame = -2
Query: 23 KPYSAPAGKGKQRL------NDERRPKGRDTPAEIVCFKCGEKGHKSNVCTKDEKKCFRC 76
KP+ P +G N RP DT +EIVC +C +KGH +N C + C+ C
Sbjct: 434 KPFQRPQNRGTSSGYSHSFGNFVPRPTQSDT-SEIVCHRCSKKGHFANRC--PDLVCWNC 264
Query: 77 GQKGHVLADCKRGDI-VCYNCNGEGHISSQCTQPKKVRTGGKVFALAGMQTTNEDRLIRG 135
+ GH DC + N + + K+ +V+ ++G ++ D LIR
Sbjct: 263 QKTGHSGKDCTNPKVEAATNAIAARRPAPAANKGKRPVASARVYTVSGAESHRADGLIRS 84
Query: 136 TCFFNSIPLIAIIDTGATHCFIALECA 162
N PL + D+GATH FI L CA
Sbjct: 83 VGSVNCKPLTILFDSGATHSFIDLACA 3
>BI418821
Length = 614
Score = 57.4 bits (137), Expect = 4e-09
Identities = 32/98 (32%), Positives = 41/98 (41%), Gaps = 15/98 (15%)
Frame = +2
Query: 26 SAPAGKGKQRLNDERRPKGRDTPAEIVCFKCGEKGHKSNVCTKDEKK--------CFRCG 77
SAP G G R ERR G C+ CG+ GH + C + C+ CG
Sbjct: 314 SAPRGFGGWR-GGERRNGGGGGGGGGGCYNCGDTGHLARDCHRSNNNGGGGGGAACYNCG 490
Query: 78 QKGHVLADCKR-------GDIVCYNCNGEGHISSQCTQ 108
GH+ DC R G CYNC GH++ C +
Sbjct: 491 DAGHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNR 604
Score = 44.3 bits (103), Expect = 4e-05
Identities = 18/52 (34%), Positives = 23/52 (43%), Gaps = 8/52 (15%)
Frame = +2
Query: 73 CFRCGQKGHVLADCKR--------GDIVCYNCNGEGHISSQCTQPKKVRTGG 116
C+ CG GH+ DC R G CYNC GH++ C + GG
Sbjct: 392 CYNCGDTGHLARDCHRSNNNGGGGGGAACYNCGDAGHLARDCNRSNNNSGGG 547
Score = 39.7 bits (91), Expect = 9e-04
Identities = 14/45 (31%), Positives = 21/45 (46%), Gaps = 7/45 (15%)
Frame = +2
Query: 53 CFKCGEKGHKSNVCTKDEKK-------CFRCGQKGHVLADCKRGD 90
C+ CG+ GH + C + C+ CG GH+ DC R +
Sbjct: 476 CYNCGDAGHLARDCNRSNNNSGGGGAGCYNCGDTGHLARDCNRSN 610
>TC11885 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, partial (26%)
Length = 555
Score = 45.8 bits (107), Expect = 1e-05
Identities = 23/69 (33%), Positives = 33/69 (47%)
Frame = +2
Query: 38 DERRPKGRDTPAEIVCFKCGEKGHKSNVCTKDEKKCFRCGQKGHVLADCKRGDIVCYNCN 97
D RR RD +C C GH + C + C CG GH+ ++C + C+NC
Sbjct: 311 DSRRGFSRDN----LCKNCKRPGHFARECP-NVAICHNCGLPGHIASECTTKSL-CWNCK 472
Query: 98 GEGHISSQC 106
GH++S C
Sbjct: 473 EPGHMASSC 499
Score = 38.5 bits (88), Expect = 0.002
Identities = 22/73 (30%), Positives = 35/73 (47%), Gaps = 5/73 (6%)
Frame = +2
Query: 24 PYSAPAGKGKQRLN---DERRPK--GRDTPAEIVCFKCGEKGHKSNVCTKDEKKCFRCGQ 78
PY + +G R N + +RP R+ P +C CG GH ++ CT + C+ C +
Sbjct: 299 PYRRDSRRGFSRDNLCKNCKRPGHFARECPNVAICHNCGLPGHIASECT-TKSLCWNCKE 475
Query: 79 KGHVLADCKRGDI 91
GH+ + C I
Sbjct: 476 PGHMASSCPNDGI 514
Score = 35.8 bits (81), Expect = 0.013
Identities = 15/35 (42%), Positives = 20/35 (56%)
Frame = +2
Query: 73 CFRCGQKGHVLADCKRGDIVCYNCNGEGHISSQCT 107
C C + GH +C I C+NC GHI+S+CT
Sbjct: 344 CKNCKRPGHFARECPNVAI-CHNCGLPGHIASECT 445
>TC9521 similar to UP|Q9FYA7 (Q9FYA7) Splicing factor RSZ33, partial (56%)
Length = 598
Score = 45.4 bits (106), Expect = 2e-05
Identities = 21/44 (47%), Positives = 23/44 (51%), Gaps = 3/44 (6%)
Frame = +1
Query: 72 KCFRCGQKGHVLADCKRGD--IVCYNCNGEGHISSQC-TQPKKV 112
+CF CG GH DCK GD CY C GHI C PKK+
Sbjct: 388 RCFNCGIDGHWARDCKAGDWKNKCYRCGERGHIEKNCKNSPKKL 519
Score = 44.7 bits (104), Expect = 3e-05
Identities = 24/65 (36%), Positives = 33/65 (49%), Gaps = 3/65 (4%)
Frame = +1
Query: 26 SAPAGKGKQRLNDERRPKGRDTP-AEIVCFKCGEKGHKSNVCTKDE--KKCFRCGQKGHV 82
S G G+ R +R GR P CF CG GH + C + KC+RCG++GH+
Sbjct: 316 SREGGGGRDR---DREYMGRGPPPGSGRCFNCGIDGHWARDCKAGDWKNKCYRCGERGHI 486
Query: 83 LADCK 87
+CK
Sbjct: 487 EKNCK 501
Score = 28.9 bits (63), Expect = 1.6
Identities = 10/23 (43%), Positives = 13/23 (56%)
Frame = +1
Query: 53 CFKCGEKGHKSNVCTKDEKKCFR 75
C++CGE+GH C KK R
Sbjct: 457 CYRCGERGHIEKNCKNSPKKLSR 525
>TC10011 similar to UP|Q9FYA7 (Q9FYA7) Splicing factor RSZ33, partial (62%)
Length = 684
Score = 45.4 bits (106), Expect = 2e-05
Identities = 23/57 (40%), Positives = 29/57 (50%), Gaps = 5/57 (8%)
Frame = +2
Query: 72 KCFRCGQKGHVLADCKRGD--IVCYNCNGEGHISSQC-TQPKK--VRTGGKVFALAG 123
+CF CG GH DCK GD CY C GH+ C PKK +T ++F+ G
Sbjct: 368 RCFNCGLDGHWARDCKAGDWKNKCYRCGDRGHVERNCKNSPKKNEWQTWKELFSFTG 538
Score = 44.7 bits (104), Expect = 3e-05
Identities = 24/59 (40%), Positives = 30/59 (50%), Gaps = 2/59 (3%)
Frame = +2
Query: 31 KGKQRLNDERRPKGRDTPAEIVCFKCGEKGHKSNVCTKDE--KKCFRCGQKGHVLADCK 87
KG R N E +G P CF CG GH + C + KC+RCG +GHV +CK
Sbjct: 308 KGGPRGNREYLGRG-PPPGSGRCFNCGLDGHWARDCKAGDWKNKCYRCGDRGHVERNCK 481
Score = 27.3 bits (59), Expect = 4.5
Identities = 8/20 (40%), Positives = 12/20 (60%)
Frame = +2
Query: 53 CFKCGEKGHKSNVCTKDEKK 72
C++CG++GH C KK
Sbjct: 437 CYRCGDRGHVERNCKNSPKK 496
>TC13053 similar to UP|Q8LF59 (Q8LF59) DNA-binding protein, partial (9%)
Length = 450
Score = 41.6 bits (96), Expect = 2e-04
Identities = 14/34 (41%), Positives = 21/34 (61%)
Frame = +3
Query: 73 CFRCGQKGHVLADCKRGDIVCYNCNGEGHISSQC 106
C C Q GH+ DC ++C+NC G GH++ +C
Sbjct: 9 CRNCQQLGHMSRDCMGPLMICHNCGGRGHLAYEC 110
Score = 36.2 bits (82), Expect = 0.010
Identities = 14/39 (35%), Positives = 19/39 (47%)
Frame = +3
Query: 51 IVCFKCGEKGHKSNVCTKDEKKCFRCGQKGHVLADCKRG 89
+VC C + GH S C C CG +GH+ +C G
Sbjct: 3 VVCRNCQQLGHMSRDCMGPLMICHNCGGRGHLAYECPSG 119
>TC17929
Length = 791
Score = 32.3 bits (72), Expect = 0.14
Identities = 11/37 (29%), Positives = 20/37 (53%)
Frame = +2
Query: 33 KQRLNDERRPKGRDTPAEIVCFKCGEKGHKSNVCTKD 69
+ ++ +E + D+ C++CGE GHK C K+
Sbjct: 2 RAQIGEETSNRPNDSKFRQTCYRCGESGHKMRNCPKE 112
Score = 28.1 bits (61), Expect = 2.6
Identities = 8/22 (36%), Positives = 15/22 (67%)
Frame = +2
Query: 67 TKDEKKCFRCGQKGHVLADCKR 88
+K + C+RCG+ GH + +C +
Sbjct: 44 SKFRQTCYRCGESGHKMRNCPK 109
>TC9306 similar to UP|Q42922 (Q42922) Annexin (Fragment), partial (59%)
Length = 662
Score = 30.0 bits (66), Expect = 0.70
Identities = 14/39 (35%), Positives = 21/39 (52%)
Frame = -3
Query: 157 IALECAYKLGLIVSDMKGEMVVETPAKGSVTTSLVCLIC 195
I EC +K ++D+ M+ G + TSLVCL+C
Sbjct: 570 IITECPHKRNQKLADVPRSMIGNVFFNGPLETSLVCLLC 454
>TC8101 similar to UP|O24074 (O24074) DNAJ-like protein, partial (90%)
Length = 1737
Score = 29.3 bits (64), Expect = 1.2
Identities = 21/70 (30%), Positives = 29/70 (41%), Gaps = 10/70 (14%)
Frame = +1
Query: 51 IVCFKCGEKGHKSNVCTKDEKKCFRCGQKGHV-------LADCKRGDIVCYNCNGEGHIS 103
++C KC KG KS +CF C G L ++ VC +C G G +
Sbjct: 583 VLCLKCKGKGSKSGTA----GRCFGCQGTGMKVIRRQIGLGMVQQMQHVCSDCRGTGEVI 750
Query: 104 SQ---CTQPK 110
S+ C Q K
Sbjct: 751 SERDRCPQCK 780
>TC9039
Length = 1218
Score = 28.9 bits (63), Expect = 1.6
Identities = 15/49 (30%), Positives = 21/49 (42%), Gaps = 6/49 (12%)
Frame = +2
Query: 26 SAPAGKGKQRLNDERRPKGRDTPA------EIVCFKCGEKGHKSNVCTK 68
S KGK++ E + + D PA + C+ C GH CTK
Sbjct: 746 STSKDKGKRKKTVEPKNEAADAPAPKKQKEDDTCYFCNVSGHMKKKCTK 892
Score = 27.7 bits (60), Expect = 3.5
Identities = 9/22 (40%), Positives = 14/22 (62%)
Frame = +2
Query: 87 KRGDIVCYNCNGEGHISSQCTQ 108
++ D CY CN GH+ +CT+
Sbjct: 827 QKEDDTCYFCNVSGHMKKKCTK 892
>BP043432
Length = 487
Score = 28.5 bits (62), Expect = 2.0
Identities = 26/90 (28%), Positives = 35/90 (38%), Gaps = 4/90 (4%)
Frame = -3
Query: 10 AAALEAVAQAVGPKPYSAPAGKGKQRLNDERRPKGRDTPAEIVCFKCGEKGHKSNVCTKD 69
A LE + Q V P S+PA Q NDE + G G K + N +D
Sbjct: 452 AIELEKLTQ-VHPSGSSSPAIGPPQEQNDEGKSIG------------GHKKKRKNETPED 312
Query: 70 ----EKKCFRCGQKGHVLADCKRGDIVCYN 95
+KK CG + C ++VC N
Sbjct: 311 LERKKKKLIECGTVADAVRTCDICNVVCNN 222
>TC18767
Length = 1004
Score = 28.1 bits (61), Expect = 2.6
Identities = 9/21 (42%), Positives = 12/21 (56%)
Frame = +2
Query: 68 KDEKKCFRCGQKGHVLADCKR 88
+D +CF CG H L +C R
Sbjct: 152 EDASRCFNCGSYNHSLRECSR 214
>BP039068
Length = 467
Score = 28.1 bits (61), Expect = 2.6
Identities = 10/21 (47%), Positives = 12/21 (56%)
Frame = +3
Query: 46 DTPAEIVCFKCGEKGHKSNVC 66
+ P + VC C E GH SN C
Sbjct: 291 NNPRQTVCMNCQETGHASNDC 353
>TC16568 similar to UP|AAR82759 (AAR82759) RE40656p (Fragment), partial
(13%)
Length = 421
Score = 28.1 bits (61), Expect = 2.6
Identities = 17/41 (41%), Positives = 22/41 (53%)
Frame = -1
Query: 58 EKGHKSNVCTKDEKKCFRCGQKGHVLADCKRGDIVCYNCNG 98
E G K V DE C + G G +LADC D + +NC+G
Sbjct: 142 EGGLKGAVGPDDEPPCCQ-GFGGGLLADC*EIDAMDWNCDG 23
>AV420911
Length = 418
Score = 27.3 bits (59), Expect = 4.5
Identities = 7/19 (36%), Positives = 13/19 (67%)
Frame = +2
Query: 69 DEKKCFRCGQKGHVLADCK 87
++ KC+ CG+ GH +C+
Sbjct: 362 EDLKCYECGEPGHFARECR 418
>TC18003 similar to PIR|T05112|T05112 splicing factor 9G8-like SR protein
RSZp22 [validated] - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (42%)
Length = 587
Score = 27.3 bits (59), Expect = 4.5
Identities = 7/19 (36%), Positives = 13/19 (67%)
Frame = +1
Query: 69 DEKKCFRCGQKGHVLADCK 87
++ KC+ CG+ GH +C+
Sbjct: 154 EDLKCYECGEPGHFARECR 210
>TC11095 similar to PIR|T05112|T05112 splicing factor 9G8-like SR protein
RSZp22 [validated] - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (89%)
Length = 912
Score = 26.9 bits (58), Expect = 5.9
Identities = 7/18 (38%), Positives = 12/18 (65%)
Frame = +1
Query: 70 EKKCFRCGQKGHVLADCK 87
+ KC+ CG+ GH +C+
Sbjct: 370 DMKCYECGEPGHFARECR 423
>TC9401 similar to UP|Q84LW7 (Q84LW7) Peptidylprolyl cis-trans isomerase ,
partial (97%)
Length = 918
Score = 26.9 bits (58), Expect = 5.9
Identities = 16/45 (35%), Positives = 24/45 (52%), Gaps = 3/45 (6%)
Frame = +1
Query: 301 PDVPPEREVEFSIDLV---PRTKPVSMAPYRMSASELAELKNQLE 342
PD+PP+ + F ++L+ PR K S+ + L ELK Q E
Sbjct: 469 PDIPPDATLVFEVELLACNPR-KGSSLGSVTEERARLEELKKQRE 600
>AV773507
Length = 496
Score = 26.6 bits (57), Expect = 7.7
Identities = 17/51 (33%), Positives = 22/51 (42%)
Frame = +2
Query: 73 CFRCGQKGHVLADCKRGDIVCYNCNGEGHISSQCTQPKKVRTGGKVFALAG 123
CF CG+ GH DC G G +SS +P+ +GG L G
Sbjct: 350 CFECGRPGHRARDCPLAGGGGGRGRGGGSLSS---RPRYGGSGGHGDRLGG 493
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.137 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,257,617
Number of Sequences: 28460
Number of extensions: 108672
Number of successful extensions: 555
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 489
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 533
length of query: 394
length of database: 4,897,600
effective HSP length: 92
effective length of query: 302
effective length of database: 2,279,280
effective search space: 688342560
effective search space used: 688342560
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 56 (26.2 bits)
Medicago: description of AC130806.14


