

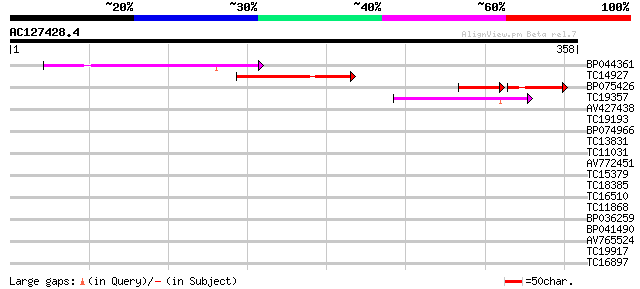
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC127428.4 + phase: 0
(358 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BP044361 84 4e-17
TC14927 weakly similar to GB|BAC16200.1|22831356|AP005486 transp... 65 2e-11
BP075426 39 5e-09
TC19357 weakly similar to UP|Q9FHY5 (Q9FHY5) Emb|CAB72466.1 (At5... 53 9e-08
AV427438 38 0.002
TC19193 similar to UP|Q9FHY5 (Q9FHY5) Emb|CAB72466.1 (At5g41980)... 35 0.025
BP074966 32 0.16
TC13831 similar to GB|AAO63315.1|28950783|BT005251 At1g23330 {Ar... 31 0.37
TC11031 homologue to UP|WR65_ARATH (Q9LP56) Probable WRKY transc... 28 3.1
AV772451 28 3.1
TC15379 UP|Q9ZPL8 (Q9ZPL8) Protein phosphatase type 2C, complete 27 4.0
TC18385 similar to UP|Q9XFL6 (Q9XFL6) Peroxidase 5, partial (38%) 27 5.3
TC16510 similar to UP|Q9FLL5 (Q9FLL5) Similarity to myosin heavy... 27 5.3
TC11868 similar to UP|Q8S9A5 (Q8S9A5) Glucosyltransferase like p... 26 9.0
BP036259 26 9.0
BP041490 26 9.0
AV765524 26 9.0
TC19917 similar to GB|AAH46895.1|28461393|BC046895 wu:fb37e02 pr... 26 9.0
TC16897 homologue to PIR|T09261|T09261 JUN kinase-activation-dom... 26 9.0
>BP044361
Length = 542
Score = 84.0 bits (206), Expect = 4e-17
Identities = 57/184 (30%), Positives = 83/184 (44%), Gaps = 45/184 (24%)
Frame = +1
Query: 22 EESYILAALLGEYATKYLCKEPCRTSELTGHAWVQEILQGNPTRCYEMFRMEKHIFHKLC 81
+E ++AA G Y L K+P R ++ E+L G+ EM RM+KH+FHKLC
Sbjct: 1 DEMELVAAAAGYYYYSCLTKQPSRRK----CEFMAEVLNGHDDFFREMLRMDKHVFHKLC 168
Query: 82 HELVEHDL-KSSKHMGVEEMVAMFLVVVGHGVGNRMIQERFQHSGETVR----------- 129
L + L + + + +EE +A+FL ++GH NR+IQERFQHSGET+
Sbjct: 169 DILRQRTLLRDTPGVMIEEQLAIFLNIIGHNERNRVIQERFQHSGETISRHFNNVLKAVK 348
Query: 130 ---------------------------------VIDGTHVSCVVSASEQPRFIGRKGYPT 156
VIDG + V A + RF +KG
Sbjct: 349 SLSREVLQPPQPTTPPEILNSARYYPYFKDCIGVIDGMQIPAHVPAKDHSRFRNKKGILA 528
Query: 157 QNIM 160
QN++
Sbjct: 529 QNVL 540
>TC14927 weakly similar to GB|BAC16200.1|22831356|AP005486 transposase-like
{Oryza sativa (japonica cultivar-group);} , partial
(11%)
Length = 539
Score = 65.1 bits (157), Expect = 2e-11
Identities = 33/75 (44%), Positives = 46/75 (61%)
Frame = +3
Query: 144 EQPRFIGRKGYPTQNIMAVCDWNMCFTFVLAGWEGTAHDARVFDKALTTANLNFPHPPQG 203
++PR+ RKG + N++ VC ++ F +VL GWEG+A D+RV AL N P G
Sbjct: 6 DKPRYRNRKGDISTNVLGVCGPDLKFIYVLPGWEGSAGDSRVLRDAL--RRQNHLQIPNG 179
Query: 204 KYYLVDSGYPTPIGY 218
KY+LVD+GY GY
Sbjct: 180 KYFLVDAGYTNGPGY 224
>BP075426
Length = 342
Score = 39.3 bits (90), Expect(2) = 5e-09
Identities = 17/29 (58%), Positives = 22/29 (75%)
Frame = -3
Query: 284 IVVATMAIHNFIRRSAEMDVDFNLYEDEN 312
I++ATM IHNFIRR + D DF +EDE+
Sbjct: 298 ILIATMTIHNFIRRKSATDSDFMHFEDED 212
Score = 37.4 bits (85), Expect(2) = 5e-09
Identities = 18/38 (47%), Positives = 29/38 (75%)
Frame = -1
Query: 315 IHHDDDHRSTNLNQSQSFNVASSSEMDHARNSIRDQII 352
+++D+D++ + ++ S VASS+EMD RNSIRDQI+
Sbjct: 207 LNNDNDNQ---VGENPSITVASSTEMDFVRNSIRDQIL 103
>TC19357 weakly similar to UP|Q9FHY5 (Q9FHY5) Emb|CAB72466.1 (At5g41980),
partial (16%)
Length = 577
Score = 52.8 bits (125), Expect = 9e-08
Identities = 35/92 (38%), Positives = 47/92 (51%), Gaps = 4/92 (4%)
Frame = +2
Query: 243 EVFNYYHSSLRCTIERTFGVWKNRFAILRSMPKFKYETQVHIVVATMAIHNFIRRSAEMD 302
E+FN+ H LR I R+F V K RF IL P++ ++ Q IV+A +HN IRR D
Sbjct: 8 ELFNHRHYFLRGAILRSFTVLKARFPILIPAPQYSFQIQRDIVIAACVLHNIIRREDRND 187
Query: 303 VDFNLY----EDENTVIHHDDDHRSTNLNQSQ 330
FN DE TV+ D + + Q Q
Sbjct: 188 WLFNSLGGTPVDELTVLDELPDVQLLSSMQEQ 283
>AV427438
Length = 182
Score = 38.1 bits (87), Expect = 0.002
Identities = 16/32 (50%), Positives = 21/32 (65%)
Frame = +2
Query: 50 TGHAWVQEILQGNPTRCYEMFRMEKHIFHKLC 81
TG +V E+L G C E FRM+K +F+KLC
Sbjct: 74 TGLKFVDEVLNGQNELCLENFRMDKDVFYKLC 169
>TC19193 similar to UP|Q9FHY5 (Q9FHY5) Emb|CAB72466.1 (At5g41980), partial
(8%)
Length = 529
Score = 34.7 bits (78), Expect = 0.025
Identities = 16/33 (48%), Positives = 22/33 (66%)
Frame = +1
Query: 279 ETQVHIVVATMAIHNFIRRSAEMDVDFNLYEDE 311
+TQV +VVA A+HNFIRR D F +Y ++
Sbjct: 4 QTQVKLVVAACALHNFIRRENPDDRLFKMYGED 102
>BP074966
Length = 169
Score = 32.0 bits (71), Expect = 0.16
Identities = 14/24 (58%), Positives = 19/24 (78%)
Frame = -3
Query: 335 ASSSEMDHARNSIRDQIIAYKLNN 358
ASS+EMD RNSIRDQ++ + +N
Sbjct: 167 ASSTEMDFVRNSIRDQLLERQCHN 96
>TC13831 similar to GB|AAO63315.1|28950783|BT005251 At1g23330 {Arabidopsis
thaliana;}, partial (40%)
Length = 567
Score = 30.8 bits (68), Expect = 0.37
Identities = 14/29 (48%), Positives = 18/29 (61%)
Frame = +1
Query: 224 CERYHLPEFRRSSGFENHNEVFNYYHSSL 252
CERYH+P S G NHN V NY ++ +
Sbjct: 466 CERYHMP---*SIGCHNHNSVNNYLNTKI 543
>TC11031 homologue to UP|WR65_ARATH (Q9LP56) Probable WRKY transcription
factor 65 (WRKY DNA-binding protein 65), partial (15%)
Length = 453
Score = 27.7 bits (60), Expect = 3.1
Identities = 20/95 (21%), Positives = 44/95 (46%), Gaps = 23/95 (24%)
Frame = +3
Query: 281 QVHIVVATMAIHNFIR--------RSAEMDVDFN---------LYEDENTVIHHDDDHRS 323
++HI++ + H + +S EMD F+ L E++ V HH ++ +
Sbjct: 3 ELHIIIIIIDHHYLVAGSRVKIQPKSQEMDSKFSNNRHRYINELLEEDAEVAHHHQENIA 182
Query: 324 TNLNQSQSFNV------ASSSEMDHARNSIRDQII 352
+ S +FN+ +S+S +R +I+ +++
Sbjct: 183 DSPPSSTAFNIDGLVTSSSTSSAKRSRRAIQKRVV 287
>AV772451
Length = 388
Score = 27.7 bits (60), Expect = 3.1
Identities = 10/25 (40%), Positives = 13/25 (52%)
Frame = +2
Query: 154 YPTQNIMAVCDWNMCFTFVLAGWEG 178
+PT NI +C W C L W+G
Sbjct: 101 HPTDNIGPICCWRECTMESLIDWQG 175
>TC15379 UP|Q9ZPL8 (Q9ZPL8) Protein phosphatase type 2C, complete
Length = 1338
Score = 27.3 bits (59), Expect = 4.0
Identities = 16/42 (38%), Positives = 24/42 (57%)
Frame = -3
Query: 48 ELTGHAWVQEILQGNPTRCYEMFRMEKHIFHKLCHELVEHDL 89
+L+GH +ILQ +PT CY++ +E+ H L HDL
Sbjct: 448 DLSGH----QILQIDPTHCYQLS*IEQQ------HNLPWHDL 353
>TC18385 similar to UP|Q9XFL6 (Q9XFL6) Peroxidase 5, partial (38%)
Length = 424
Score = 26.9 bits (58), Expect = 5.3
Identities = 8/16 (50%), Positives = 10/16 (62%)
Frame = +2
Query: 162 VCDWNMCFTFVLAGWE 177
+C W CF F+ GWE
Sbjct: 209 LCQWR*CFHFIRPGWE 256
>TC16510 similar to UP|Q9FLL5 (Q9FLL5) Similarity to myosin heavy chain,
partial (6%)
Length = 489
Score = 26.9 bits (58), Expect = 5.3
Identities = 12/30 (40%), Positives = 16/30 (53%)
Frame = -2
Query: 255 TIERTFGVWKNRFAILRSMPKFKYETQVHI 284
TI F +WK +F R K+K T +HI
Sbjct: 329 TIRIAFSIWKKKFKAWRRKRKYKNCTIMHI 240
>TC11868 similar to UP|Q8S9A5 (Q8S9A5) Glucosyltransferase like protein
(Fragment), partial (25%)
Length = 485
Score = 26.2 bits (56), Expect = 9.0
Identities = 9/15 (60%), Positives = 11/15 (73%)
Frame = -1
Query: 240 NHNEVFNYYHSSLRC 254
NHN+ NY+HSS C
Sbjct: 440 NHNKHMNYFHSSFFC 396
>BP036259
Length = 532
Score = 26.2 bits (56), Expect = 9.0
Identities = 10/20 (50%), Positives = 12/20 (60%)
Frame = -3
Query: 2 NFWKEQLEHNTQEEEDDDTF 21
N W E E ++EEDDD F
Sbjct: 176 NIWLEGHEREEEDEEDDDDF 117
>BP041490
Length = 545
Score = 26.2 bits (56), Expect = 9.0
Identities = 9/29 (31%), Positives = 19/29 (65%)
Frame = -2
Query: 52 HAWVQEILQGNPTRCYEMFRMEKHIFHKL 80
H +V ++++ RC++MF +++ I H L
Sbjct: 364 HCFVMQVMRSTTRRCFQMF-LQRSITHNL 281
>AV765524
Length = 600
Score = 26.2 bits (56), Expect = 9.0
Identities = 9/31 (29%), Positives = 17/31 (54%)
Frame = +3
Query: 62 NPTRCYEMFRMEKHIFHKLCHELVEHDLKSS 92
+PT+CY + + ++ K+C H L+ S
Sbjct: 219 SPTQCYTLCTKKGWLYPKICRNKSNHSLQKS 311
>TC19917 similar to GB|AAH46895.1|28461393|BC046895 wu:fb37e02 protein
{Danio rerio;} , partial (8%)
Length = 491
Score = 26.2 bits (56), Expect = 9.0
Identities = 12/40 (30%), Positives = 23/40 (57%)
Frame = +3
Query: 89 LKSSKHMGVEEMVAMFLVVVGHGVGNRMIQERFQHSGETV 128
L ++++ E VAMFL ++ H + R++ + S ET+
Sbjct: 366 LVRTRNVPTIEAVAMFLHILAHNLKYRVVHFTYYRSKETI 485
>TC16897 homologue to PIR|T09261|T09261 JUN kinase-activation-domain-binding
protein homolog - alfalfa {Medicago
sativa;}, partial (37%)
Length = 505
Score = 26.2 bits (56), Expect = 9.0
Identities = 14/52 (26%), Positives = 29/52 (54%), Gaps = 1/52 (1%)
Frame = -1
Query: 292 HNFIRRSAEMDVDFNLYEDENTVIHHDDD-HRSTNLNQSQSFNVASSSEMDH 342
H FIR S +D F+ + ++ IH DD +L+++ F+ A+ + +++
Sbjct: 478 HVFIRISLGIDTGFSAFNGQSESIHDDDRIGVGFSLHEAHDFDGAAGARVNN 323
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.322 0.136 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,283,405
Number of Sequences: 28460
Number of extensions: 115472
Number of successful extensions: 556
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 550
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 554
length of query: 358
length of database: 4,897,600
effective HSP length: 91
effective length of query: 267
effective length of database: 2,307,740
effective search space: 616166580
effective search space used: 616166580
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 56 (26.2 bits)
Medicago: description of AC127428.4


